Strikingly Different Roles of SARS-CoV-2 Fusion Peptides Uncovered by Neutron Scattering
- PMID: 35157798
- PMCID: PMC8862744
- DOI: 10.1021/jacs.1c09856
Strikingly Different Roles of SARS-CoV-2 Fusion Peptides Uncovered by Neutron Scattering
Abstract
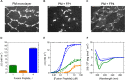
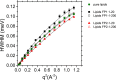
Coronavirus disease-2019 (COVID-19), a potentially lethal respiratory illness caused by the coronavirus SARS-CoV-2, emerged in the end of 2019 and has since spread aggressively across the globe. A thorough understanding of the molecular mechanisms of cellular infection by coronaviruses is therefore of utmost importance. A critical stage in infection is the fusion between viral and host membranes. Here, we present a detailed investigation of the role of selected SARS-CoV-2 Spike fusion peptides, and the influence of calcium and cholesterol, in this fusion process. Structural information from specular neutron reflectometry and small angle neutron scattering, complemented by dynamics information from quasi-elastic and spin-echo neutron spectroscopy, revealed strikingly different functions encoded in the Spike fusion domain. Calcium drives the N-terminal of the Spike fusion domain to fully cross the host plasma membrane. Removing calcium, however, reorients the peptide back to the lipid leaflet closest to the virus, leading to significant changes in lipid fluidity and rigidity. In conjunction with other regions of the fusion domain, which are also positioned to bridge and dehydrate viral and host membranes, the molecular events leading to cell entry by SARS-CoV-2 are proposed.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Li Q.; Wu J.; Nie J.; Zhang L.; Hao H.; Liu S.; Zhao C.; Zhang Q.; Liu H.; Nie L.; Qin H.; Wang M.; Lu Q.; Li X.; Sun Q.; Liu J.; Zhang L.; Li X.; Huang W.; Wang Y. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182 (5), 1284–1294. 10.1016/j.cell.2020.07.012. - DOI - PMC - PubMed
-
- Daly J. L.; Simonetti B.; Klein K.; Chen K. E.; Williamson M. K.; Anton-Plagaro C.; Shoemark D. K.; Simon-Gracia L.; Bauer M.; Hollandi R.; Greber U. F.; Horvath P.; Sessions R. B.; Helenius A.; Hiscox J. A.; Teesalu T.; Matthews D. A.; Davidson A. D.; Collins B. M.; Cullen P. J.; Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370 (6518), 861–865. 10.1126/science.abd3072. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

