Structural Variability of Lipoarabinomannan Modulates Innate Immune Responses within Infected Alveolar Epithelial Cells
- PMID: 35159170
- PMCID: PMC8834380
- DOI: 10.3390/cells11030361
Structural Variability of Lipoarabinomannan Modulates Innate Immune Responses within Infected Alveolar Epithelial Cells
Abstract
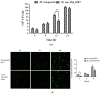
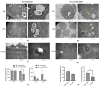
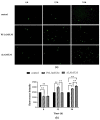
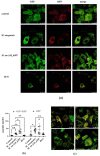
Mycobacterium tuberculosis (M. tb) is an intracellular pathogen persisting in phagosomes that has the ability to escape host immune surveillance causing tuberculosis (TB). Lipoarabinomannan (LAM), as a glycolipid, is one of the complex outermost components of the mycobacterial cell envelope and plays a critical role in modulating host responses during M. tb infection. Different species within the Mycobacterium genus exhibit distinct LAM structures and elicit diverse innate immune responses. However, little is known about the mechanisms. In this study, we first constructed a LAM-truncated mutant with fewer arabinofuranose (Araf) residues named M. sm-ΔM_6387 (Mycobacterium smegmatis arabinosyltransferase EmbC gene knockout strain). It exhibited some prominent cell wall defects, including tardiness of mycobacterial migration, loss of acid-fast staining, and increased cell wall permeability. Within alveolar epithelial cells (A549) infected by M. sm-ΔM_6387, the uptake rate was lower, phagosomes with bacterial degradation appeared, and microtubule-associated protein light chain 3 (LC3) recruitment was enhanced compared to wild type Mycobacteriumsmegmatis (M. smegmatis). We further confirmed that the variability in the removal capability of M. sm-ΔM_6387 resulted from host cell responses rather than the changes in the mycobacterial cell envelope. Moreover, we found that M. sm-ΔM_6387 or its glycolipid extracts significantly induced expression changes in some genes related to innate immune responses, including Toll-like receptor 2 (TLR2), class A scavenger receptor (SR-A), Rubicon, LC3, tumor necrosis factor alpha (TNF-α), Bcl-2, and Bax. Therefore, our studies suggest that nonpathogenic M. smegmatis can deposit LC3 on phagosomal membranes, and the decrease in the quantity of Araf residues for LAM molecules not only impacts mycobacterial cell wall integrity but also enhances host defense responses against the intracellular pathogens and decreases phagocytosis of host cells.
Keywords: EmbC; LC3-associated phagocytosis; Mycobacterium smegmatis; alveolar epithelial cells; lipoarabinomannan.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Mutations in the essential arabinosyltransferase EmbC lead to alterations in Mycobacterium tuberculosis lipoarabinomannan.J Biol Chem. 2014 Dec 19;289(51):35172-81. doi: 10.1074/jbc.M114.583112. Epub 2014 Oct 28. J Biol Chem. 2014. PMID: 25352598 Free PMC article.
-
Critical roles for lipomannan and lipoarabinomannan in cell wall integrity of mycobacteria and pathogenesis of tuberculosis.mBio. 2013 Feb 19;4(1):e00472-12. doi: 10.1128/mBio.00472-12. mBio. 2013. PMID: 23422411 Free PMC article.
-
The critical role of embC in Mycobacterium tuberculosis.J Bacteriol. 2008 Jun;190(12):4335-41. doi: 10.1128/JB.01825-07. Epub 2008 Apr 18. J Bacteriol. 2008. PMID: 18424526 Free PMC article.
-
Lipoarabinomannan, and its related glycolipids, induce divergent and opposing immune responses to Mycobacterium tuberculosis depending on structural diversity and experimental variations.Tuberculosis (Edinb). 2016 Jan;96:120-30. doi: 10.1016/j.tube.2015.09.005. Epub 2015 Oct 28. Tuberculosis (Edinb). 2016. PMID: 26586646 Review.
-
Polysaccharides and glycolipids of Mycobacterium tuberculosis and their induced immune responses.Scand J Immunol. 2023 May;97(5):e13261. doi: 10.1111/sji.13261. Epub 2023 Mar 6. Scand J Immunol. 2023. PMID: 39008002 Review.
Cited by
-
In Silico Targeting and Immunological Profiling of PpiA in Mycobacterium tuberculosis: A Computational Approach.Pathogens. 2025 Apr 9;14(4):370. doi: 10.3390/pathogens14040370. Pathogens. 2025. PMID: 40333132 Free PMC article.
-
Clinical application of the urinary lipoarabinomannan (AIMLAM) test in PLHIV with TB.AIDS Res Ther. 2025 Jun 7;22(1):59. doi: 10.1186/s12981-025-00754-4. AIDS Res Ther. 2025. PMID: 40483460 Free PMC article.
-
Mycobacterial lipoarabinomannan negatively interferes with macrophage responses to Aspergillus fumigatus in-vitro.bioRxiv [Preprint]. 2024 Nov 20:2024.11.18.623945. doi: 10.1101/2024.11.18.623945. bioRxiv. 2024. PMID: 39605324 Free PMC article. Preprint.
References
-
- World Health Organization . Global Tuberculosis Report 2021. World Health Organization; Geneva, Switzerland: 2021.
-
- Chakaya J., Khan M., Ntoumi F., Aklillu E., Fatima R., Mwaba P., Kapata N., Mfinanga S., Hasnain S.E., Katoto P.D.M.C., et al. Global Tuberculosis Report 2020—Reflections on the Global TB burden, treatment and prevention efforts. Int. J. Infect. Dis. 2021;113:S7–S12. doi: 10.1016/j.ijid.2021.02.107. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

