Histone Chaperone Nrp1 Mutation Affects the Acetylation of H3K56 in Tetrahymena thermophila
- PMID: 35159218
- PMCID: PMC8833950
- DOI: 10.3390/cells11030408
Histone Chaperone Nrp1 Mutation Affects the Acetylation of H3K56 in Tetrahymena thermophila
Abstract
Histone modification and nucleosome assembly are mainly regulated by various histone-modifying enzymes and chaperones. The roles of histone-modification enzymes have been well analyzed, but the molecular mechanism of histone chaperones in histone modification and nucleosome assembly is incompletely understood. We previously found that the histone chaperone Nrp1 is localized in the micronucleus (MIC) and the macronucleus (MAC) and involved in the chromatin stability and nuclear division of Tetrahymena thermophila. In the present work, we found that truncated C-terminal mutant HA-Nrp1TrC abnormally localizes in the cytoplasm. The truncated-signal-peptide mutants HA-Nrp1TrNLS1 and HA-Nrp1TrNLS2 are localized in the MIC and MAC. Overexpression of Nrp1TrNLS1 inhibited cellular proliferation and disrupted micronuclear mitosis during the vegetative growth stage. During sexual development, Nrp1TrNLS1 overexpression led to abnormal bouquet structures and meiosis arrest. Furthermore, Histone H3 was not transported into the nucleus; instead, it formed an abnormal speckled cytoplastic distribution in the Nrp1TrNLS1 mutants. The acetylation level of H3K56 in the mutants also decreased, leading to significant changes in the transcription of the genome of the Nrp1TrNLS1 mutants. The histone chaperone Nrp1 regulates the H3 nuclear import and acetylation modification of H3K56 and affects chromatin stability and genome transcription in Tetrahymena.
Keywords: Tetrahymena thermophila; acetylation of H3K56; genome transcription; histone chaperone NRP1; mutation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

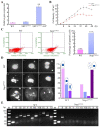
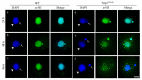


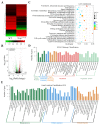
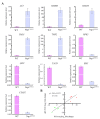
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

