Use of CRISPR/Cas9-Based Gene Editing to Simultaneously Mutate Multiple Homologous Genes Required for Pollen Development and Male Fertility in Maize
- PMID: 35159251
- PMCID: PMC8834288
- DOI: 10.3390/cells11030439
Use of CRISPR/Cas9-Based Gene Editing to Simultaneously Mutate Multiple Homologous Genes Required for Pollen Development and Male Fertility in Maize
Abstract
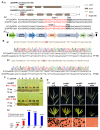
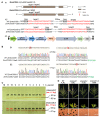
Male sterility represents an important trait for hybrid breeding and seed production in crops. Although the genes required for male fertility have been widely studied and characterized in many plant species, most of them are single genic male-sterility (GMS) genes. To investigate the role of multiple homologous genes in anther and pollen developments of maize, we established the CRISPR/Cas9-based gene editing method to simultaneously mutate the homologs in several putative GMS gene families. By using the integrated strategies of multi-gene editing vectors, maize genetic transformation, mutation-site analysis of T0 and F1 plants, and genotyping and phenotyping of F2 progenies, we further confirmed gene functions of every member in ZmTGA9-1/-2/-3 family, and identified the functions of ZmDFR1, ZmDFR2, ZmACOS5-1, and ZmACOS5-2 in controlling maize male fertility. Single and double homozygous gene mutants of ZmTGA9-1/-2/-3 did not affect anther and pollen development, while triple homozygous gene mutant resulted in complete male sterility. Two single-gene mutants of ZmDFR1/2 displayed partial male sterility, but the double-gene mutant showed complete male sterility. Additionally, only the ZmACOS5-2 single gene was required for anther and pollen development, while ZmACOS5-1 had no effect on male fertility. Our results show that the CRISPR/Cas9 gene editing system is a highly efficient and convenient tool for identifying multiple homologous GMS genes. These findings enrich GMS genes and mutant resources for breeding of maize GMS lines and promote deep understanding of the gene family underlying pollen development and male fertility in maize.
Keywords: CRISPR/Cas9; gene editing; genic male sterility; maize; multiple homologous genes; pollen development.
Conflict of interest statement
The authors declare no conflict of interest. Beijing Solidwill Sci-Tech Co., Ltd., the Beijing Engineering Laboratory, and Beijing International Science and Technology Cooperation Base provided the research platform, instruments, and funds for this study; thus, the company is one of the signed units of this paper.
Figures


Similar articles
-
CRISPR/Cas9-based discovery of maize transcription factors regulating male sterility and their functional conservation in plants.Plant Biotechnol J. 2021 Sep;19(9):1769-1784. doi: 10.1111/pbi.13590. Epub 2021 May 4. Plant Biotechnol J. 2021. PMID: 33772993 Free PMC article.
-
CRISPR/Cas9-based genome editing of 14 lipid metabolic genes reveals a sporopollenin metabolon ZmPKSB-ZmTKPR1-1/-2 required for pollen exine formation in maize.Plant Biotechnol J. 2024 Jan;22(1):216-232. doi: 10.1111/pbi.14181. Epub 2023 Oct 4. Plant Biotechnol J. 2024. PMID: 37792967 Free PMC article.
-
Map-Based Cloning, Phylogenetic, and Microsynteny Analyses of ZmMs20 Gene Regulating Male Fertility in Maize.Int J Mol Sci. 2019 Mar 20;20(6):1411. doi: 10.3390/ijms20061411. Int J Mol Sci. 2019. PMID: 30897816 Free PMC article.
-
Maize Genic Male-Sterility Genes and Their Applications in Hybrid Breeding: Progress and Perspectives.Mol Plant. 2019 Mar 4;12(3):321-342. doi: 10.1016/j.molp.2019.01.014. Epub 2019 Jan 26. Mol Plant. 2019. PMID: 30690174 Review.
-
Genetic Structure and Molecular Mechanisms Underlying the Formation of Tassel, Anther, and Pollen in the Male Inflorescence of Maize (Zea mays L.).Cells. 2022 May 26;11(11):1753. doi: 10.3390/cells11111753. Cells. 2022. PMID: 35681448 Free PMC article. Review.
Cited by
-
Genetic structure and molecular mechanism underlying the stalk lodging traits in maize (Zea mays L.).Comput Struct Biotechnol J. 2022 Dec 21;21:485-494. doi: 10.1016/j.csbj.2022.12.037. eCollection 2023. Comput Struct Biotechnol J. 2022. PMID: 36618981 Free PMC article.
-
Mechanisms, Machinery, and Dynamics of Chromosome Segregation in Zea mays.Genes (Basel). 2024 Dec 16;15(12):1606. doi: 10.3390/genes15121606. Genes (Basel). 2024. PMID: 39766873 Free PMC article. Review.
-
Efficient mutagenesis and genotyping of maize inbreds using biolistics, multiplex CRISPR/Cas9 editing, and Indel-Selective PCR.Plant Methods. 2025 Mar 25;21(1):43. doi: 10.1186/s13007-025-01365-w. Plant Methods. 2025. PMID: 40128730 Free PMC article.
-
Effect of high temperature on pollen grains and yield in economically important crops: a review.Planta. 2025 May 16;261(6):141. doi: 10.1007/s00425-025-04714-0. Planta. 2025. PMID: 40374974 Review.
-
CRISPR-Cas technology opens a new era for the creation of novel maize germplasms.Front Plant Sci. 2022 Dec 16;13:1049803. doi: 10.3389/fpls.2022.1049803. eCollection 2022. Front Plant Sci. 2022. PMID: 36589095 Free PMC article. Review.
References
-
- Virmani S., Ilyas-Ahmed M. Environment-sensitive genic male sterility (EGMS) in crops. Adv. Agron. 2001;72:139–195. doi: 10.1016/S0065-2113(01)72013-5. - DOI
-
- Wu Y., Fox T.W., Trimnell M.R., Wang L., Xu R.j., Cigan A.M., Huffman G.A., Garnaat C.W., Hershey H., Albertsen M.C. Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 2016;14:1046–1054. doi: 10.1111/pbi.12477. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

