Evidence of Adult Features and Functions of Hepatocytes Differentiated from Human Induced Pluripotent Stem Cells and Self-Organized as Organoids
- PMID: 35159346
- PMCID: PMC8834365
- DOI: 10.3390/cells11030537
Evidence of Adult Features and Functions of Hepatocytes Differentiated from Human Induced Pluripotent Stem Cells and Self-Organized as Organoids
Abstract
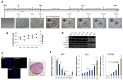
Background: Human-induced pluripotent stem cell-derived hepatocytes (iHeps) have been shown to have considerable potential in liver diseases, toxicity, and pharmacological studies. However, there is a growing need to obtain iHeps that are truly similar to primary adult hepatocytes in terms of morphological features and functions. We generated such human iHeps, self-assembled as organoids (iHep-Orgs).
Methods: iPSC-derived hepatoblasts were self-assembled into spheroids and differentiated into mature hepatocytes modulating final step of differentiation.
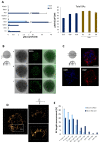
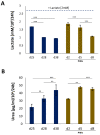
Results: In about four weeks of culture, the albumin secretion levels and the complete disappearance of α-fetoprotein from iHep-Orgs suggested the acquisition of a greater degree of maturation than those previously reported. The expression of apical transporters and bile acid secretion evidenced the acquisition of complex hepatocyte polarity as well as the development of a functional and well-defined bile canalicular network confirmed by computational analysis. Activities recorded for CYP450, UGT1A1, and alcohol dehydrogenase, response to hormonal stimulation, and glucose metabolism were also remarkable. Finally, iHep-Orgs displayed a considerable ability to detoxify pathological concentrations of lactate and ammonia.
Conclusions: With features similar to those of primary adult hepatocytes, the iHep-Orgs thus produced could be considered as a valuable tool for the development and optimization of preclinical and clinical applications.
Keywords: HLCs; bile acids; bile canaliculi network; detoxification; hepatic mature functions; hiPSC-derived hepatocytes; hiPSCs; liver organoids; metabolism; self-assembling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
The Potential Clinical Use of Stem/Progenitor Cells and Organoids in Liver Diseases.Cells. 2022 Apr 21;11(9):1410. doi: 10.3390/cells11091410. Cells. 2022. PMID: 35563716 Free PMC article. Review.
-
Microencapsulated Hepatocytes Differentiated from Human Induced Pluripotent Stem Cells: Optimizing 3D Culture for Tissue Engineering Applications.Cells. 2023 Mar 10;12(6):865. doi: 10.3390/cells12060865. Cells. 2023. PMID: 36980206 Free PMC article.
-
Engineered microtissues to model the effects of dynamic heterotypic cell signaling on iPSC-derived human hepatocyte maturation.Acta Biomater. 2025 May 1;197:135-151. doi: 10.1016/j.actbio.2025.03.020. Epub 2025 Mar 13. Acta Biomater. 2025. PMID: 40089127
-
3-D culture and endothelial cells improve maturity of human pluripotent stem cell-derived hepatocytes.Acta Biomater. 2019 Sep 1;95:371-381. doi: 10.1016/j.actbio.2019.07.047. Epub 2019 Jul 27. Acta Biomater. 2019. PMID: 31362140
-
Human Pluripotent Stem Cell-Derived Hepatocyte-Like Cells and Organoids for Liver Disease and Therapy.Int J Mol Sci. 2021 Sep 28;22(19):10471. doi: 10.3390/ijms221910471. Int J Mol Sci. 2021. PMID: 34638810 Free PMC article. Review.
Cited by
-
Cell-based approaches for the mechanistic understanding of drug-induced cholestatic liver injury.Arch Toxicol. 2025 Jun;99(6):2243-2260. doi: 10.1007/s00204-025-04016-0. Epub 2025 Mar 11. Arch Toxicol. 2025. PMID: 40064699 Free PMC article. Review.
-
Guiding Hepatic Differentiation of Pluripotent Stem Cells Using 3D Microfluidic Co-Cultures with Human Hepatocytes.Cells. 2023 Aug 1;12(15):1982. doi: 10.3390/cells12151982. Cells. 2023. PMID: 37566061 Free PMC article.
-
The Potential Clinical Use of Stem/Progenitor Cells and Organoids in Liver Diseases.Cells. 2022 Apr 21;11(9):1410. doi: 10.3390/cells11091410. Cells. 2022. PMID: 35563716 Free PMC article. Review.
-
Microencapsulated Hepatocytes Differentiated from Human Induced Pluripotent Stem Cells: Optimizing 3D Culture for Tissue Engineering Applications.Cells. 2023 Mar 10;12(6):865. doi: 10.3390/cells12060865. Cells. 2023. PMID: 36980206 Free PMC article.
-
Stem Cells and Organoids: A Paradigm Shift in Preclinical Models Toward Personalized Medicine.Pharmaceuticals (Basel). 2025 Jul 1;18(7):992. doi: 10.3390/ph18070992. Pharmaceuticals (Basel). 2025. PMID: 40732281 Free PMC article. Review.
References
-
- Holmgren G., Ulfenborg B., Asplund A., Toet K., Andersson C.X., Hammarstedt A., Hanemaaijer R., Küppers-Munther B., Synnergren J. Characterization of Human Induced Pluripotent Stem Cell-Derived Hepatocytes with Mature Features and Potential for Modeling Metabolic Diseases. Int. J. Mol. Sci. 2020;21:469. doi: 10.3390/ijms21020469. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

