Interactome of Arabidopsis Thaliana
- PMID: 35161331
- PMCID: PMC8838453
- DOI: 10.3390/plants11030350
Interactome of Arabidopsis Thaliana
Abstract
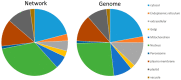
More than 95,000 protein-protein interactions of Arabidopsis thaliana have been published and deposited in databases. This dataset was supplemented by approximately 900 additional interactions, which were identified in the literature from the years 2002-2021. These protein-protein interactions were used as the basis for a Cytoscape network and were supplemented with data on subcellular localization, gene ontologies, biochemical properties and co-expression. The resulting network has been exemplarily applied in unraveling the PPI-network of the plant vacuolar proton-translocating ATPase (V-ATPase), which was selected due to its central importance for the plant cell. In particular, it is involved in cellular pH homeostasis, providing proton motive force necessary for transport processes, trafficking of proteins and, thereby, cell wall synthesis. The data points to regulation taking place on multiple levels: (a) a phosphorylation-dependent regulation by 14-3-3 proteins and by kinases such as WNK8 and NDPK1a, (b) an energy-dependent regulation via HXK1 and the glucose receptor RGS1 and (c) a Ca2+-dependent regulation by SOS2 and IDQ6. The known importance of V-ATPase for cell wall synthesis is supported by its interactions with several proteins involved in cell wall synthesis. The resulting network was further analyzed for (experimental) biases and was found to be enriched in nuclear, cytosolic and plasma membrane proteins but depleted in extracellular and mitochondrial proteins, in comparison to the entity of protein-coding genes. Among the processes and functions, proteins involved in transcription were highly abundant in the network. Subnetworks were extracted for organelles, processes and protein families. The degree of representation of organelles and processes reveals limitations and advantages in the current knowledge of protein-protein interactions, which have been mainly caused by a high number of database entries being contributed by only a few publications with highly specific motivations and methodologies that favor, for instance, interactions in the cytosol and the nucleus.
Keywords: Arabidopsis thaliana; Cytoscape; interactome; protein–protein interaction.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

