Comparison of Magnoliaceae Plastomes: Adding Neotropical Magnolia to the Discussion
- PMID: 35161429
- PMCID: PMC8838774
- DOI: 10.3390/plants11030448
Comparison of Magnoliaceae Plastomes: Adding Neotropical Magnolia to the Discussion
Abstract
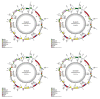
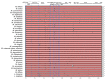
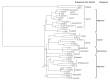
Chloroplast genomes are considered to be highly conserved. Nevertheless, differences in their sequences are an important source of phylogenetically informative data. Chloroplast genomes are increasingly applied in evolutionary studies of angiosperms, including Magnoliaceae. Recent studies have focused on resolving the previously debated classification of the family using a phylogenomic approach and chloroplast genome data. However, most Neotropical clades and recently described species have not yet been included in molecular studies. We performed sequencing, assembly, and annotation of 15 chloroplast genomes from Neotropical Magnoliaceae species. We compared the newly assembled chloroplast genomes with 22 chloroplast genomes from across the family, including representatives from each genus and section. Family-wide, the chloroplast genomes presented a length of about 160 kb. The gene content in all species was constant, with 145 genes. The intergenic regions showed a higher level of nucleotide diversity than the coding regions. Differences were higher among genera than within genera. The phylogenetic analysis in Magnolia showed two main clades and corroborated that the current infrageneric classification does not represent natural groups. Although chloroplast genomes are highly conserved in Magnoliaceae, the high level of diversity of the intergenic regions still resulted in an important source of phylogenetically informative data, even for closely related taxa.
Keywords: chloroplast assembly; comparative genomics; complete chloroplast genome; phylogenomics; whole genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures












References
-
- Cooper G. The Cell: A Molecular Approach. ASM Press; New York, NY, USA: 2007. Chloroplast and Other Plastids; p. 820.
-
- Grivet D., Heinze B., Vendramin G.G., Petit R.J. Genome walking with consensus primers: Application to the large single copy region of chloroplast DNA. Mol. Ecol. Notes. 2001;1:345–349. doi: 10.1046/j.1471-8278.2001.00107.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

