Linear Combination Properties of the Phasor Space in Fluorescence Imaging
- PMID: 35161742
- PMCID: PMC8840623
- DOI: 10.3390/s22030999
Linear Combination Properties of the Phasor Space in Fluorescence Imaging
Abstract
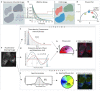
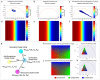
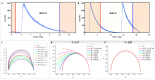
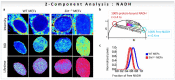
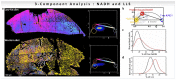
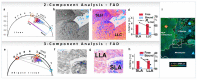
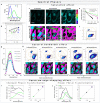
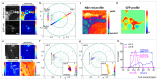
The phasor approach to fluorescence lifetime imaging, and more recently hyperspectral fluorescence imaging, has increased the use of these techniques, and improved the ease and intuitiveness of the data analysis. The fit-free nature of the phasor plots increases the speed of the analysis and reduces the dimensionality, optimization of data handling and storage. The reciprocity principle between the real and imaginary space-where the phasor and the pixel that the phasor originated from are linked and can be converted from one another-has helped the expansion of this method. The phasor coordinates calculated from a pixel, where multiple fluorescent species are present, depends on the phasor positions of those components. The relative positions are governed by the linear combination properties of the phasor space. According to this principle, the phasor position of a pixel with multiple components lies inside the polygon whose vertices are occupied by the phasor positions of these individual components and the distance between the image phasor to any of the vertices is inversely proportional to the fractional intensity contribution of that component to the total fluorescence from that image pixel. The higher the fractional intensity contribution of a vertex, the closer is the resultant phasor. The linear additivity in the phasor space can be exploited to obtain the fractional intensity contribution from multiple species and quantify their contribution. This review details the various mathematical models that can be used to obtain two/three/four components from phasor space with known phasor signatures and then how to obtain both the fractional intensities and phasor positions without any prior knowledge of either, assuming they are mono-exponential in nature. We note that other than for blind components, there are no restrictions on the type of the decay or their phasor positions for linear combinations to be valid-and they are applicable to complicated fluorescence lifetime decays from components with intensity decays described by multi-exponentials.
Keywords: FLIM; fractional intensity; hyperspectral imaging; linear combination of phasor; model-free; multidimensional phasor plot; multiple component analysis; phasor; spectral phasor.
Conflict of interest statement
The authors do not have any conflict of interest.
Figures






Similar articles
-
Blind Resolution of Lifetime Components in Individual Pixels of Fluorescence Lifetime Images Using the Phasor Approach.J Phys Chem B. 2020 Nov 12;124(45):10126-10137. doi: 10.1021/acs.jpcb.0c06946. Epub 2020 Nov 3. J Phys Chem B. 2020. PMID: 33140960 Free PMC article.
-
Resolution of 4 components in the same pixel in FLIM images using the phasor approach.Methods Appl Fluoresc. 2020 Apr 15;8(3):035001. doi: 10.1088/2050-6120/ab8570. Methods Appl Fluoresc. 2020. PMID: 32235070
-
Fit-free analysis of fluorescence lifetime imaging data using the phasor approach.Nat Protoc. 2018 Sep;13(9):1979-2004. doi: 10.1038/s41596-018-0026-5. Nat Protoc. 2018. PMID: 30190551
-
The Phasor Plot: A Universal Circle to Advance Fluorescence Lifetime Analysis and Interpretation.Annu Rev Biophys. 2021 May 6;50:575-593. doi: 10.1146/annurev-biophys-062920-063631. Annu Rev Biophys. 2021. PMID: 33957055 Review.
-
Label-free fluorescence microscopy: revisiting the opportunities with autofluorescent molecules and harmonic generations as biosensors and biomarkers for quantitative biology.Biophys Rev. 2023 Jun 26;15(4):709-719. doi: 10.1007/s12551-023-01083-4. eCollection 2023 Aug. Biophys Rev. 2023. PMID: 37681086 Free PMC article. Review.
Cited by
-
FLUTE: A Python GUI for interactive phasor analysis of FLIM data.Biol Imaging. 2023 Nov 6;3:e21. doi: 10.1017/S2633903X23000211. eCollection 2023. Biol Imaging. 2023. PMID: 38487690 Free PMC article.
-
Lipid packing and cholesterol content regulate membrane wetting and remodeling by biomolecular condensates.Nat Commun. 2025 Mar 20;16(1):2756. doi: 10.1038/s41467-025-57985-2. Nat Commun. 2025. PMID: 40113768 Free PMC article.
-
Oral N-acetylcysteine decreases IFN-γ production and ameliorates ischemia-reperfusion injury in steatotic livers.Front Immunol. 2022 Sep 5;13:898799. doi: 10.3389/fimmu.2022.898799. eCollection 2022. Front Immunol. 2022. PMID: 36148239 Free PMC article.
-
Unravelling the different components of nonphotochemical quenching using a novel analytical pipeline.New Phytol. 2025 Jan;245(2):625-636. doi: 10.1111/nph.20271. Epub 2024 Nov 15. New Phytol. 2025. PMID: 39545639 Free PMC article.
-
Fluorescence phasor analysis: basic principles and biophysical applications.Biophys Rev. 2025 Mar 7;17(2):395-408. doi: 10.1007/s12551-025-01293-y. eCollection 2025 Apr. Biophys Rev. 2025. PMID: 40376409 Review.
References
-
- Weber G. Resolution of the Fluorescence Lifetimes in A Heterogeneous System by Phase and Modulation Measurements. J. Phys. Chem. 1981;85:949–953. doi: 10.1021/j150608a006. - DOI
-
- Stringari C., Cinquin A., Cinquin O., Digman M.A., Donovan P.J., Gratton E. Phasor Approach to Fluorescence Lifetime Microscopy Distinguishes Different Metabolic States of Germ Cells in A Live Tissue. Proc. Natl. Acad. Sci. USA. 2011;108:13582–13587. doi: 10.1073/pnas.1108161108. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

