Identification and Allele Combination Analysis of Rice Grain Shape-Related Genes by Genome-Wide Association Study
- PMID: 35162989
- PMCID: PMC8835367
- DOI: 10.3390/ijms23031065
Identification and Allele Combination Analysis of Rice Grain Shape-Related Genes by Genome-Wide Association Study
Abstract
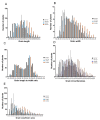
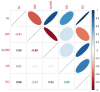
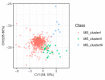
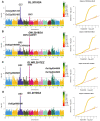
Grain shape is an important agronomic character of rice, which affects the appearance, processing, and the edible quality. Screening and identifying more new genes associated with grain shape is beneficial to further understanding the genetic basis of grain shape and provides more gene resources for genetic breeding. This study has a natural population containing 623 indica rice cultivars. Genome-wide association studies/GWAS of several traits related to grain shape (grain length/GL, grain width/GW, grain length to width ratio/GLWR, grain circumferences/GC, and grain size/grain area/GS) were conducted by combining phenotypic data from four environments and the second-generation resequencing data, which have identified 39 important Quantitative trait locus/QTLs. We analyzed the 39 QTLs using three methods: gene-based association analysis, haplotype analysis, and functional annotation and identified three cloned genes (GS3, GW5, OsDER1) and seven new candidate genes in the candidate interval. At the same time, to effectively utilize the genes in the grain shape-related gene bank, we have also analyzed the allelic combinations of the three cloned genes. Finally, the extreme allele combination corresponding to each trait was found through statistical analysis. This study's novel candidate genes and allele combinations will provide a valuable reference for future breeding work.
Keywords: GWAS; allele combination analysis; gene-based association analysis; grain shape; haplotype analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Redoña E.D., Mackill D.J. Quantitative trait locus analysis for rice panicle and grain characteristics. Theor. Appl. Genet. 1998;96:957–963. doi: 10.1007/s001220050826. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

