CRISPR Editing Enables Consequential Tag-Activated MicroRNA-Mediated Endogene Deactivation
- PMID: 35163006
- PMCID: PMC8834719
- DOI: 10.3390/ijms23031082
CRISPR Editing Enables Consequential Tag-Activated MicroRNA-Mediated Endogene Deactivation
Abstract
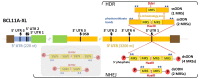
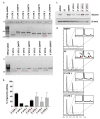
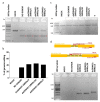
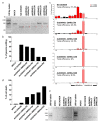
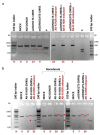
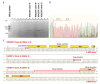
Molecular therapies and functional studies greatly benefit from spatial and temporal precision of genetic intervention. We therefore conceived and explored tag-activated microRNA (miRNA)-mediated endogene deactivation (TAMED) as a research tool and potential lineage-specific therapy. For proof of principle, we aimed to deactivate γ-globin repressor BCL11A in erythroid cells by tagging the 3' untranslated region (UTR) of BCL11A with miRNA recognition sites (MRSs) for the abundant erythromiR miR-451a. To this end, we employed nucleofection of CRISPR/Cas9 ribonucleoprotein (RNP) particles alongside double- or single-stranded oligodeoxynucleotides for, respectively, non-homologous-end-joining (NHEJ)- or homology-directed-repair (HDR)-mediated MRS insertion. NHEJ-based tagging was imprecise and inefficient (≤6%) and uniformly produced knock-in- and indel-containing MRS tags, whereas HDR-based tagging was more efficient (≤18%), but toxic for longer donors encoding concatenated and thus potentially more efficient MRS tags. Isolation of clones for robust HEK293T cells tagged with a homozygous quadruple MRS resulted in 25% spontaneous reduction in BCL11A and up to 36% reduction after transfection with an miR-451a mimic. Isolation of clones for human umbilical cord blood-derived erythroid progenitor-2 (HUDEP-2) cells tagged with single or double MRS allowed detection of albeit weak γ-globin induction. Our study demonstrates suitability of TAMED for physiologically relevant modulation of gene expression and its unsuitability for therapeutic application in its current form.
Keywords: BCL11A; CD34+ cells; HUDEP-2 cells; cell/tissue-type-specific gene therapy; erythroid-specific; gene tagging; hemoglobinopathies; γ-globin induction.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
CRISPR/Cas9-based multiplex genome editing of BCL11A and HBG efficiently induces fetal hemoglobin expression.Eur J Pharmacol. 2022 Mar 5;918:174788. doi: 10.1016/j.ejphar.2022.174788. Epub 2022 Jan 28. Eur J Pharmacol. 2022. PMID: 35093321
-
Transcriptional Repressor BCL11A in Erythroid Cells.Adv Exp Med Biol. 2024;1459:199-215. doi: 10.1007/978-3-031-62731-6_9. Adv Exp Med Biol. 2024. PMID: 39017845 Review.
-
Dynamics and competition of CRISPR-Cas9 ribonucleoproteins and AAV donor-mediated NHEJ, MMEJ and HDR editing.Nucleic Acids Res. 2021 Jan 25;49(2):969-985. doi: 10.1093/nar/gkaa1251. Nucleic Acids Res. 2021. PMID: 33398341 Free PMC article.
-
A Cas9-transcription factor fusion protein enhances homology-directed repair efficiency.J Biol Chem. 2021 Jan-Jun;296:100525. doi: 10.1016/j.jbc.2021.100525. Epub 2021 Mar 6. J Biol Chem. 2021. PMID: 33689695 Free PMC article.
-
Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish.Methods. 2017 May 15;121-122:77-85. doi: 10.1016/j.ymeth.2017.03.005. Epub 2017 Mar 12. Methods. 2017. PMID: 28300641 Review.
Cited by
-
In search of an ideal template for therapeutic genome editing: A review of current developments for structure optimization.Front Genome Ed. 2023 Feb 22;5:1068637. doi: 10.3389/fgeed.2023.1068637. eCollection 2023. Front Genome Ed. 2023. PMID: 36911237 Free PMC article. Review.
-
Progress and harmonization of gene editing to treat human diseases: Proceeding of COST Action CA21113 GenE-HumDi.Mol Ther Nucleic Acids. 2023 Oct 29;34:102066. doi: 10.1016/j.omtn.2023.102066. eCollection 2023 Dec 12. Mol Ther Nucleic Acids. 2023. PMID: 38034032 Free PMC article. Review.
-
High-efficiency editing in hematopoietic stem cells and the HUDEP-2 cell line based on in vitro mRNA synthesis.Front Genome Ed. 2023 Mar 8;5:1141618. doi: 10.3389/fgeed.2023.1141618. eCollection 2023. Front Genome Ed. 2023. PMID: 36969374 Free PMC article.
References
-
- Sakamoto K., Rädler P.D., Wehde B.L., Triplett A.A., Shrestha H., Ferraiuolo R.M., Amari F., Coppola V., Klinakis A., Efstratiadis A., et al. Efficient tissue-type specific expression of target genes in a tetracycline-controlled manner from the ubiquitously active Eef1a1 locus. Sci. Rep. 2020;10 doi: 10.1038/s41598-019-57052-z. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

