Detecting Bacterial-Human Lateral Gene Transfer in Chronic Lymphocytic Leukemia
- PMID: 35163016
- PMCID: PMC8835664
- DOI: 10.3390/ijms23031094
Detecting Bacterial-Human Lateral Gene Transfer in Chronic Lymphocytic Leukemia
Abstract
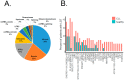
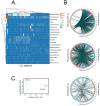
Chronic lymphocytic leukemia (CLL) is a very common and mostly incurable B-cell malignancy. Recent studies revealed high interpatient mutational heterogeneity and worsened therapy response and survival of patients with complex genomic aberrations. In line with this, a better understanding of the underlying mechanisms of specific genetic aberrations would reveal new prognostic factors and possible therapeutic targets. It is known that chromosomal rearrangements including DNA insertions often play a role during carcinogenesis. Recently it was reported that bacteria (microbiome)-human lateral gene transfer occurs in somatic cells and is enriched in cancer samples. To further investigate this mechanism in CLL, we analyzed paired-end RNA sequencing data of 45 CLL patients and 9 healthy donors, in which we particularly searched for bacterial DNA integrations into the human somatic genome. Applying the Burrows-Wheeler aligner (BWA) first on a human genome and then on bacterial genome references, we differentiated between sequencing reads mapping to the human genome, to the microbiome or to bacterial integrations into the human genome. Our results indicate that CLL samples featured bacterial DNA integrations more frequently (approx. two-fold) compared to normal samples, which corroborates the latest findings in other cancer entities. Moreover, we determined common integration sites and recurrent integrated bacterial transcripts. Finally, we investigated the contribution of bacterial integrations to oncogenesis and disease progression.
Keywords: CLL; bacterial integrations; lateral gene transfer (LGT).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Visentin A., Facco M., Gurrieri C., Pagnin E., Martini V., Imbergamo S., Frezzato F., Trimarco V., Severin F., Raggi F., et al. Prognostic and Predictive Effect of IGHV Mutational Status and Load in Chronic Lymphocytic Leukemia: Focus on FCR and BR Treatments. Clin. Lymphoma Myeloma Leuk. 2019;19:678–685.e4. doi: 10.1016/j.clml.2019.03.002. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

