The Current State of Chromatin Immunoprecipitation (ChIP) from FFPE Tissues
- PMID: 35163027
- PMCID: PMC8834906
- DOI: 10.3390/ijms23031103
The Current State of Chromatin Immunoprecipitation (ChIP) from FFPE Tissues
Abstract
Cancer cells accumulate epigenomic aberrations that contribute to cancer initiation and progression by altering both the genomic stability and the expression of genes. The awareness of such alterations could improve our understanding of cancer dynamics and the identification of new therapeutic strategies and biomarkers to refine tumor classification and treatment. Formalin fixation and paraffin embedding (FFPE) is the gold standard to preserve both tissue integrity and organization, and, in the last decades, a huge number of biological samples have been archived all over the world following this procedure. Recently, new chromatin immunoprecipitation (ChIP) techniques have been developed to allow the analysis of histone post-translational modifications (PTMs) and transcription factor (TF) distribution in FFPE tissues. The application of ChIP to genome-wide chromatin studies using real archival samples represents an unprecedented opportunity to conduct retrospective clinical studies thanks to the possibility of accessing large cohorts of samples and their associated diagnostic records. However, although recent attempts to standardize have been made, fixation and storage conditions of clinical specimens are still extremely variable and can affect the success of chromatin studies. The procedures introduced in the last few years dealt with this problem proponing successful strategies to obtain high-resolution ChIP profiles from FFPE archival samples. In this review, we compare the different FFPE-ChIP techniques, highlighting their strengths, limitations, common features, and peculiarities, as well as pitfalls and caveats related to ChIP studies in FFPE samples, in order to facilitate their application.
Keywords: FFPE tissues; archival samples; cancer epigenetics; chromatin; chromatin immunoprecipitation (ChIP).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

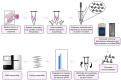
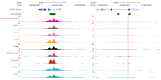
References
-
- Kadoch C. Structure and function of ATP-dependent chromatin remodeling complexes in human cancer. Blood. 2019;134((Suppl. S1)):SCI-48. doi: 10.1182/blood-2019-121033. - DOI
-
- Gomez A.P., Ilter D., Low V., Rosenzweig A., Shen Z.J., Schild T., Rivas M.A., Er E.E., McNally D.R., Mutvei A.P., et al. Dynamic incorporation of histone H3 variants into chromatin is essential for acquisition of aggressive traits and metastatic colonization. Cancer Cell. 2019;36:402–417. doi: 10.1016/j.ccell.2019.08.006. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

