Sesquiterpene Lactones Potentiate Olaparib-Induced DNA Damage in p53 Wildtype Cancer Cells
- PMID: 35163037
- PMCID: PMC8835362
- DOI: 10.3390/ijms23031116
Sesquiterpene Lactones Potentiate Olaparib-Induced DNA Damage in p53 Wildtype Cancer Cells
Abstract
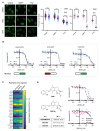
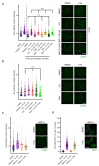
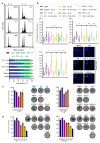
Despite notable advances in utilising PARP inhibitor monotherapy, many cancers are not PARP inhibitor-sensitive or develop treatment resistance. In this work, we show that the two structurally-related sesquiterpene lactones, a 2-bromobenzyloxy derivative of dehydrosantonin (BdS) and alantolactone (ATL) sensitise p53 wildtype, homologous recombination-proficient cancer cells to low-dose treatment with the PARP inhibitor, olaparib. Exposure to combination treatments of olaparib with BdS or ATL induces cell-cycle changes, chromosomal instability, as well as considerable increases in nuclear area. Mechanistically, we uncover that mitotic errors likely depend on oxidative stress elicited by the electrophilic lactone warheads and olaparib-mediated PARP-trapping, culminating in replication stress. Combination treatments exhibit moderately synergistic effects on cell survival, probably attenuated by a p53-mediated, protective cell-cycle arrest in the G2 cell-cycle phase. Indeed, using a WEE1 inhibitor, AZD1775, to inhibit the G2/M cell-cycle checkpoint further decreased cell survival. Around half of all cancers diagnosed retain p53 functionality, and this proportion could be expected to increase with improved diagnostic approaches in the clinic. Utilising sublethal oxidative stress to sensitise p53 wildtype, homologous recombination-proficient cancer cells to low-dose PARP-trapping could therefore serve as the basis for future research into the treatment of cancers currently refractory to PARP inhibition.
Keywords: 2-bromobenzyloxy derivative of dehydrosantonin (BdS); DNA replication stress; PARP inhibitor (PARPi); alantolactone (ATL); cancer; olaparib; reactive oxygen species (ROS); sesquiterpene lactones.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Augmented antitumor activity by olaparib plus AZD1775 in gastric cancer through disrupting DNA damage repair pathways and DNA damage checkpoint.J Exp Clin Cancer Res. 2018 Jun 28;37(1):129. doi: 10.1186/s13046-018-0790-7. J Exp Clin Cancer Res. 2018. PMID: 29954437 Free PMC article.
-
Synergistic lethality between PARP-trapping and alantolactone-induced oxidative DNA damage in homologous recombination-proficient cancer cells.Oncogene. 2020 Apr;39(14):2905-2920. doi: 10.1038/s41388-020-1191-x. Epub 2020 Feb 6. Oncogene. 2020. PMID: 32029902 Free PMC article.
-
PARP1 Trapping and DNA Replication Stress Enhance Radiosensitization with Combined WEE1 and PARP Inhibitors.Mol Cancer Res. 2018 Feb;16(2):222-232. doi: 10.1158/1541-7786.MCR-17-0455. Epub 2017 Nov 13. Mol Cancer Res. 2018. PMID: 29133592 Free PMC article.
-
A comprehensive review of anticancer mechanisms of action of Alantolactone.Biomed Pharmacother. 2021 Apr;136:111231. doi: 10.1016/j.biopha.2021.111231. Epub 2021 Jan 14. Biomed Pharmacother. 2021. PMID: 33454597 Review.
-
Green nanotech paradigm for enhancing sesquiterpene lactone therapeutics in cancer.Biomed Pharmacother. 2024 Apr;173:116426. doi: 10.1016/j.biopha.2024.116426. Epub 2024 Mar 11. Biomed Pharmacother. 2024. PMID: 38471274 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

