Genome-Wide Identification and Characterization of the Soybean DEAD-Box Gene Family and Expression Response to Rhizobia
- PMID: 35163041
- PMCID: PMC8835661
- DOI: 10.3390/ijms23031120
Genome-Wide Identification and Characterization of the Soybean DEAD-Box Gene Family and Expression Response to Rhizobia
Abstract
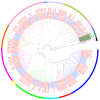
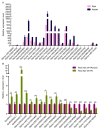
DEAD-box proteins are a large family of RNA helicases that play important roles in almost all cellular RNA processes in model plants. However, little is known about this family of proteins in crops such as soybean. Here, we identified 80 DEAD-box family genes in the Glycine max (soybean) genome. These DEAD-box genes were distributed on 19 chromosomes, and some genes were clustered together. The majority of DEAD-box family proteins were highly conserved in Arabidopsis and soybean, but Glyma.08G231300 and Glyma.14G115100 were specific to soybean. The promoters of these DEAD-box genes share cis-acting elements involved in plant responses to MeJA, salicylic acid (SA), low temperature and biotic as well as abiotic stresses; interestingly, half of the genes contain nodulation-related cis elements in their promoters. Microarray data analysis revealed that the DEAD-box genes were differentially expressed in the root and nodule. Notably, 31 genes were induced by rhizobia and/or were highly expressed in the nodule. Real-time quantitative PCR analysis validated the expression patterns of some DEAD-box genes, and among them, Glyma.08G231300 and Glyma.14G115100 were induced by rhizobia in root hair. Thus, we provide a comprehensive view of the DEAD-box family genes in soybean and highlight the crucial role of these genes in symbiotic nodulation.
Keywords: DEAD-box RNA helicases; Glycine max; nodulation; phylogenetic analysis; rhizobia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Genome-Wide Identification and Characterization of the Soybean Snf2 Gene Family and Expression Response to Rhizobia.Int J Mol Sci. 2023 Apr 14;24(8):7250. doi: 10.3390/ijms24087250. Int J Mol Sci. 2023. PMID: 37108411 Free PMC article.
-
The promoters of two isoflavone synthase genes respond differentially to nodulation and defense signals in transgenic soybean roots.Plant Mol Biol. 2004 Mar;54(5):623-39. doi: 10.1023/B:PLAN.0000040814.28507.35. Plant Mol Biol. 2004. PMID: 15356384
-
A soybean acyl carrier protein, GmACP, is important for root nodule symbiosis.Mol Plant Microbe Interact. 2014 May;27(5):415-23. doi: 10.1094/MPMI-09-13-0269-R. Mol Plant Microbe Interact. 2014. PMID: 24400939
-
Genome-wide identification, characterisation and expression profile analysis of DEAD-box family genes in sweet potato wild ancestor Ipomoea trifida under abiotic stresses.Genes Genomics. 2020 Mar;42(3):325-335. doi: 10.1007/s13258-019-00910-x. Epub 2020 Jan 1. Genes Genomics. 2020. PMID: 31894476
-
Genome-wide identification, evolution, and expression analysis of carbonic anhydrases genes in soybean (Glycine max).Funct Integr Genomics. 2023 Jan 14;23(1):37. doi: 10.1007/s10142-023-00966-9. Funct Integr Genomics. 2023. PMID: 36639600 Review.
Cited by
-
ZmHPAT2 Regulates Maize Growth and Development and Mycorrhizal Symbiosis.Plants (Basel). 2025 May 11;14(10):1438. doi: 10.3390/plants14101438. Plants (Basel). 2025. PMID: 40431003 Free PMC article.
-
Genome-wide systematic survey and analysis of the RNA helicase gene family and their response to abiotic stress in sweetpotato.BMC Plant Biol. 2024 Mar 16;24(1):193. doi: 10.1186/s12870-024-04824-z. BMC Plant Biol. 2024. PMID: 38493089 Free PMC article.
-
LkARF7 and LkARF19 overexpression promote adventitious root formation in a heterologous poplar model by positively regulating LkBBM1.Commun Biol. 2023 Apr 5;6(1):372. doi: 10.1038/s42003-023-04731-3. Commun Biol. 2023. PMID: 37020138 Free PMC article.
-
Genome-Wide Identification and Characterization of the Soybean Snf2 Gene Family and Expression Response to Rhizobia.Int J Mol Sci. 2023 Apr 14;24(8):7250. doi: 10.3390/ijms24087250. Int J Mol Sci. 2023. PMID: 37108411 Free PMC article.
-
Functions and mechanisms of RNA helicases in plants.J Exp Bot. 2023 Apr 9;74(7):2295-2310. doi: 10.1093/jxb/erac462. J Exp Bot. 2023. PMID: 36416783 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- 31961133029/National Natural Science Foundation of China
- 31730066/National Natural Science Foundation of China
- 2016YFA0500503/The National Key Research and Development Program of China
- 2020CFA008/The Natural Science Foundation of Hubei Province
- 2015RC014/Huazhong Agricultural University Scientific & Technological Self-innovation Foundation
LinkOut - more resources
Full Text Sources

