The Promoter of the Immune-Modulating Gene TIR-Containing Protein C of the Uropathogenic Escherichia coli Strain CFT073 Reacts to the Pathogen's Environment
- PMID: 35163072
- PMCID: PMC8835471
- DOI: 10.3390/ijms23031148
The Promoter of the Immune-Modulating Gene TIR-Containing Protein C of the Uropathogenic Escherichia coli Strain CFT073 Reacts to the Pathogen's Environment
Abstract
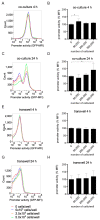
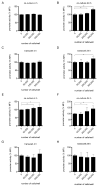
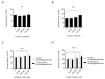
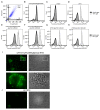
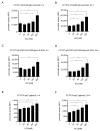
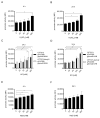
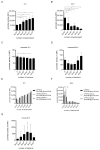
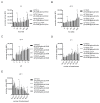
The TIR-containing protein C (TcpC) of the uropathogenic Escherichia coli strain CFT073 modulates innate immunity by interfering with the Toll-like receptor and NALP3 inflammasome signaling cascade. During a urinary tract infection the pathogen encounters epithelial and innate immune cells and replicates by several orders of magnitude. We therefore analyzed whether these cell types and also the density of the pathogen would induce the recently defined promoter of the CFT073 tcpC gene to, in time, dampen innate immune responses. Using reporter constructs we found that the uroepithelial cell line T24/83 and the monocytic cell line THP-1 induced the tcpC promoter. Differentiation of monocytic THP-1 cells to macrophages increased their potential to switch on the promoter. Cell-associated CFT073 displayed the highest promoter activity. Since potassium represents the most abundant intracellular ion and is secreted to induce the NLRP3 inflammasome, we tested its ability to activate the tcpC promoter. Potassium induced the promoter with high efficiency. Sodium, which is enriched in the renal cortex generating an antibacterial hypersalinity, also induced the tcpC promoter. Finally, the bacterial density modulated the tcpC promoter activity. In the search for promoter-regulating proteins, we found that the DNA-binding protein H-NS dampens the promoter activity. Taken together, different cell types and salts, present in the kidney, are able to induce the tcpC promoter and might explain the mechanism of TcpC induction during a kidney infection with uropathogenic E. coli strains.
Keywords: TcpC; bacterial density; host cell; potassium; sodium; uropathogenic Escherichia coli.
Conflict of interest statement
Authors have no conflict of interest to declare.
Figures
Similar articles
-
Uropathogenic Escherichia coli strain CFT073 disrupts NLRP3 inflammasome activation.J Clin Invest. 2016 Jul 1;126(7):2425-36. doi: 10.1172/JCI81916. Epub 2016 May 23. J Clin Invest. 2016. PMID: 27214553 Free PMC article.
-
Inhibition of TIR domain signaling by TcpC: MyD88-dependent and independent effects on Escherichia coli virulence.PLoS Pathog. 2010 Sep 23;6(9):e1001120. doi: 10.1371/journal.ppat.1001120. PLoS Pathog. 2010. PMID: 20886104 Free PMC article.
-
Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073.Infect Immun. 2012 Jan;80(1):321-32. doi: 10.1128/IAI.05322-11. Epub 2011 Sep 19. Infect Immun. 2012. PMID: 21930758 Free PMC article.
-
[Progress in TcpC research].Zhejiang Da Xue Xue Bao Yi Xue Ban. 2013 Sep;42(5):481-5. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2013. PMID: 24167126 Review. Chinese.
-
Classical chaperone-usher (CU) adhesive fimbriome: uropathogenic Escherichia coli (UPEC) and urinary tract infections (UTIs).Folia Microbiol (Praha). 2020 Feb;65(1):45-65. doi: 10.1007/s12223-019-00719-x. Epub 2019 Jun 5. Folia Microbiol (Praha). 2020. PMID: 31165977 Review.
Cited by
-
TcpC Inhibits M1 but Promotes M2 Macrophage Polarization via Regulation of the MAPK/NF-κB and Akt/STAT6 Pathways in Urinary Tract Infection.Cells. 2022 Aug 28;11(17):2674. doi: 10.3390/cells11172674. Cells. 2022. PMID: 36078080 Free PMC article.
-
Pseudomonas aeruginosa senses and responds to epithelial potassium flux via Kdp operon to promote biofilm.PLoS Pathog. 2024 May 31;20(5):e1011453. doi: 10.1371/journal.ppat.1011453. eCollection 2024 May. PLoS Pathog. 2024. PMID: 38820569 Free PMC article.
-
Molecular Mechanisms of Bacterial Communication and Their Biocontrol.Int J Mol Sci. 2024 May 16;25(10):5443. doi: 10.3390/ijms25105443. Int J Mol Sci. 2024. PMID: 38791481 Free PMC article.
References
-
- Mobley H.L., Green D.M., Trifillis A.L., Johnson D.E., Chippendale G.R., Lockatell C.V., Jones B.D., Warren J.W. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: Role of hemolysin in some strains. Infect. Immun. 1990;58:1281–1289. doi: 10.1128/iai.58.5.1281-1289.1990. - DOI - PMC - PubMed
-
- Welch R.A., Burland V., Plunkett G., III, Redford P., Roesch P., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. - DOI - PMC - PubMed
-
- Cirl C., Wieser A., Yadav M., Duerr S., Schubert S., Fischer H., Stappert D., Wantia N., Rodriguez N., Wagner H., et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 2008;14:399–406. doi: 10.1038/nm1734. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

