Spheroid Culture Differentially Affects Cancer Cell Sensitivity to Drugs in Melanoma and RCC Models
- PMID: 35163092
- PMCID: PMC8835769
- DOI: 10.3390/ijms23031166
Spheroid Culture Differentially Affects Cancer Cell Sensitivity to Drugs in Melanoma and RCC Models
Abstract
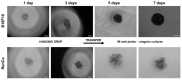
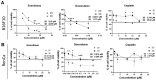
2D culture as a model for drug testing often turns to be clinically futile. Therefore, 3D cultures (3Ds) show potential to better model responses to drugs observed in vivo. In preliminary studies, using melanoma (B16F10) and renal (RenCa) cancer, we confirmed that 3Ds better mimics the tumor microenvironment. Here, we evaluated how the proposed 3D mode of culture affects tumor cell susceptibility to anti-cancer drugs, which have distinct mechanisms of action (everolimus, doxorubicin, cisplatin). Melanoma spheroids showed higher resistance to all used drugs, as compared to 2D. In an RCC model, such modulation was only observed for doxorubicin treatment. As drug distribution was not affected by the 3D shape, we assessed the expression of MDR1 and mTor. Upregulation of MDR1 in RCC spheroids was observed, in contrast to melanoma. In both models, mTor expression was not affected by the 3D cultures. By NGS, 10 genes related with metabolism of xenobiotics by cytochrome p450 were deregulated in renal cancer spheroids; 9 of them were later confirmed in the melanoma model. The differences between 3D models and classical 2D cultures point to the potential to uncover new non-canonical mechanisms to explain drug resistance set by the tumor in its microenvironment.
Keywords: 3D; RCC; cisplatin; cytochrome; doxorubicin; drug-resistance; everolimus; glutathione-s-transferases; melanoma; spheroids.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Metastatic renal cell carcinoma cells growing in 3D on poly‑D‑lysine or laminin present a stem‑like phenotype and drug resistance.Oncol Rep. 2019 Nov;42(5):1878-1892. doi: 10.3892/or.2019.7321. Epub 2019 Sep 18. Oncol Rep. 2019. PMID: 31545459 Free PMC article.
-
Three-dimensional (3D) culture of bone-derived human 786-O renal cell carcinoma retains relevant clinical characteristics of bone metastases.Cancer Lett. 2015 Aug 28;365(1):89-95. doi: 10.1016/j.canlet.2015.05.019. Epub 2015 May 21. Cancer Lett. 2015. PMID: 26004343 Free PMC article.
-
AlgiMatrix™ based 3D cell culture system as an in-vitro tumor model for anticancer studies.PLoS One. 2013;8(1):e53708. doi: 10.1371/journal.pone.0053708. Epub 2013 Jan 18. PLoS One. 2013. PMID: 23349734 Free PMC article.
-
Sunitinib, sorafenib and mTOR inhibitors in renal cancer.J BUON. 2007 Sep;12 Suppl 1:S151-62. J BUON. 2007. PMID: 17935273 Review.
-
MicroRNAs Associated with Von Hippel-Lindau Pathway in Renal Cell Carcinoma: A Comprehensive Review.Int J Mol Sci. 2017 Nov 22;18(11):2495. doi: 10.3390/ijms18112495. Int J Mol Sci. 2017. PMID: 29165391 Free PMC article. Review.
Cited by
-
c-Met+ Cytotoxic T Lymphocytes Exhibit Enhanced Cytotoxicity in Mice and Humans In Vitro Tumor Models.Biomedicines. 2023 Nov 23;11(12):3123. doi: 10.3390/biomedicines11123123. Biomedicines. 2023. PMID: 38137344 Free PMC article.
-
Comparative Analysis of the Effect of the BRAF Inhibitor Dabrafenib in 2D and 3D Cell Culture Models of Human Metastatic Melanoma Cells.In Vivo. 2024 Jul-Aug;38(4):1579-1593. doi: 10.21873/invivo.13608. In Vivo. 2024. PMID: 38936891 Free PMC article.
-
Current Research Trends in the Application of In Vitro Three-Dimensional Models of Liver Cells.Pharmaceutics. 2022 Dec 24;15(1):54. doi: 10.3390/pharmaceutics15010054. Pharmaceutics. 2022. PMID: 36678683 Free PMC article. Review.
-
Single-Cell Analysis Reveals that Vitamin C Inhibits Bone Metastasis of Renal Cancer via Cell Cycle Arrest and Microenvironment Remodeling.Adv Sci (Weinh). 2025 Aug;12(30):e01011. doi: 10.1002/advs.202501011. Epub 2025 May 28. Adv Sci (Weinh). 2025. PMID: 40433925 Free PMC article.
-
Assessment of in vitro anti-skin aging activities of Phyllanthus indofischeri Bennet extracts for dermatological and aesthetic applications.Sci Rep. 2023 Oct 31;13(1):18661. doi: 10.1038/s41598-023-45434-3. Sci Rep. 2023. PMID: 37907639 Free PMC article.
References
-
- Szenajch J., Szabelska-Beręsewicz A., Świercz A., Zyprych-Walczak J., Siatkowski I., Góralski M., Synowiec A., Handschuh L. Transcriptome Remodeling in Gradual Development of Inverse Resistance between Paclitaxel and Cisplatin in Ovarian Cancer Cells. Int. J. Mol. Sci. 2020;21:9218. doi: 10.3390/ijms21239218. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

