Amino Acids and IGF1 Regulation of Fish Muscle Growth Revealed by Transcriptome and microRNAome Integrative Analyses of Pacu (Piaractus mesopotamicus) Myotubes
- PMID: 35163102
- PMCID: PMC8835699
- DOI: 10.3390/ijms23031180
Amino Acids and IGF1 Regulation of Fish Muscle Growth Revealed by Transcriptome and microRNAome Integrative Analyses of Pacu (Piaractus mesopotamicus) Myotubes
Abstract
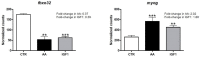
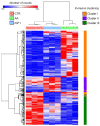
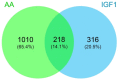
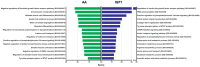
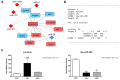
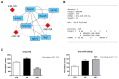
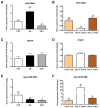
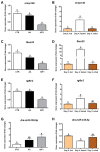
Amino acids (AA) and IGF1 have been demonstrated to play essential roles in protein synthesis and fish muscle growth. The myoblast cell culture is useful for studying muscle regulation, and omics data have contributed enormously to understanding its molecular biology. However, to our knowledge, no study has performed the large-scale sequencing of fish-cultured muscle cells stimulated with pro-growth signals. In this work, we obtained the transcriptome and microRNAome of pacu (Piaractus mesopotamicus)-cultured myotubes treated with AA or IGF1. We identified 1228 and 534 genes differentially expressed by AA and IGF1. An enrichment analysis showed that AA treatment induced chromosomal changes, mitosis, and muscle differentiation, while IGF1 modulated IGF/PI3K signaling, metabolic alteration, and matrix structure. In addition, potential molecular markers were similarly modulated by both treatments. Muscle-miRNAs (miR-1, -133, -206 and -499) were up-regulated, especially in AA samples, and we identified molecular networks with omics integration. Two pairs of genes and miRNAs demonstrated a high-level relationship, and involvement in myogenesis and muscle growth: marcksb and miR-29b in AA, and mmp14b and miR-338-5p in IGF1. Our work helps to elucidate fish muscle physiology and metabolism, highlights potential molecular markers, and creates a perspective for improvements in aquaculture and in in vitro meat production.
Keywords: IGF1; amino acids; cell culture; muscle growth; omics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Johnston I.A. Genetic and Environmental Determinants of Muscle Growth Patterns. In: Johnston I.A., editor. Muscle Development and Growth. Academic Press; Cambridge, MA, USA: 2001. pp. 141–186.
-
- Sänger A.M., Stoiber W. Muscle fiber diversity and plasticity. In: Johnston I.A., editor. Muscle Development and Growth. Academic Press; Cambridge, MA, USA: 2001. pp. 187–250. Fish Physiology.
MeSH terms
Substances
Grants and funding
- #2018/24575-6/São Paulo Research Foundation
- #2018/26428-0/São Paulo Research Foundation
- #306678/2021-7/National Council for Scientific and Technological Development
- #403305/2021-7/National Council for Scientific and Technological Development
- #88887.482392/2020-00/Coordenação de Aperfeicoamento de Pessoal de Nível Superior
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

