The Epigenetic Role of Vitamin C in Neurodevelopment
- PMID: 35163133
- PMCID: PMC8836017
- DOI: 10.3390/ijms23031208
The Epigenetic Role of Vitamin C in Neurodevelopment
Abstract
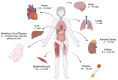
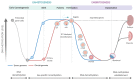
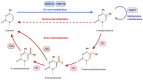
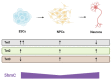
The maternal diet during pregnancy is a key determinant of offspring health. Early studies have linked poor maternal nutrition during gestation with a propensity for the development of chronic conditions in offspring. These conditions include cardiovascular disease, type 2 diabetes and even compromised mental health. While multiple factors may contribute to these outcomes, disturbed epigenetic programming during early development is one potential biological mechanism. The epigenome is programmed primarily in utero, and during this time, the developing fetus is highly susceptible to environmental factors such as nutritional insults. During neurodevelopment, epigenetic programming coordinates the formation of primitive central nervous system structures, neurogenesis, and neuroplasticity. Dysregulated epigenetic programming has been implicated in the aetiology of several neurodevelopmental disorders such as Tatton-Brown-Rahman syndrome. Accordingly, there is great interest in determining how maternal nutrient availability in pregnancy might affect the epigenetic status of offspring, and how such influences may present phenotypically. In recent years, a number of epigenetic enzymes that are active during embryonic development have been found to require vitamin C as a cofactor. These enzymes include the ten-eleven translocation methylcytosine dioxygenases (TETs) and the Jumonji C domain-containing histone lysine demethylases that catalyse the oxidative removal of methyl groups on cytosines and histone lysine residues, respectively. These enzymes are integral to epigenetic regulation and have fundamental roles in cellular differentiation, the maintenance of pluripotency and development. The dependence of these enzymes on vitamin C for optimal catalytic activity illustrates a potentially critical contribution of the nutrient during mammalian development. These insights also highlight a potential risk associated with vitamin C insufficiency during pregnancy. The link between vitamin C insufficiency and development is particularly apparent in the context of neurodevelopment and high vitamin C concentrations in the brain are indicative of important functional requirements in this organ. Accordingly, this review considers the evidence for the potential impact of maternal vitamin C status on neurodevelopmental epigenetics.
Keywords: TET enzymes; ascorbate; epigenetic programming; maternal nutrition; neurodevelopment; ten-eleven translocation methylcytosine dioxygenases; vitamin C.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Pearson J.F., Pullar J.M., Wilson R., Spittlehouse J.K., Vissers M.C.M., Skidmore P.M.L., Willis J., Cameron V.A., Carr A.C. Vitamin C Status Correlates with Markers of Metabolic and Cognitive Health in 50-Year-Olds: Findings of the CHALICE Cohort Study. Nutrients. 2017;9:831. doi: 10.3390/nu9080831. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

