Unravelling Mechanisms of Doxorubicin-Induced Toxicity in 3D Human Intestinal Organoids
- PMID: 35163210
- PMCID: PMC8836276
- DOI: 10.3390/ijms23031286
Unravelling Mechanisms of Doxorubicin-Induced Toxicity in 3D Human Intestinal Organoids
Abstract
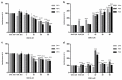
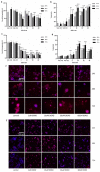
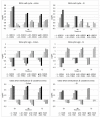
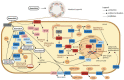
Doxorubicin is widely used in the treatment of different cancers, and its side effects can be severe in many tissues, including the intestines. Symptoms such as diarrhoea and abdominal pain caused by intestinal inflammation lead to the interruption of chemotherapy. Nevertheless, the molecular mechanisms associated with doxorubicin intestinal toxicity have been poorly explored. This study aims to investigate such mechanisms by exposing 3D small intestine and colon organoids to doxorubicin and to evaluate transcriptomic responses in relation to viability and apoptosis as physiological endpoints. The in vitro concentrations and dosing regimens of doxorubicin were selected based on physiologically based pharmacokinetic model simulations of treatment regimens recommended for cancer patients. Cytotoxicity and cell morphology were evaluated as well as gene expression and biological pathways affected by doxorubicin. In both types of organoids, cell cycle, the p53 signalling pathway, and oxidative stress were the most affected pathways. However, significant differences between colon and SI organoids were evident, particularly in essential metabolic pathways. Short time-series expression miner was used to further explore temporal changes in gene profiles, which identified distinct tissue responses. Finally, in silico proteomics revealed important proteins involved in doxorubicin metabolism and cellular processes that were in line with the transcriptomic responses, including cell cycle and senescence, transport of molecules, and mitochondria impairment. This study provides new insight into doxorubicin-induced effects on the gene expression levels in the intestines. Currently, we are exploring the potential use of these data in establishing quantitative systems toxicology models for the prediction of drug-induced gastrointestinal toxicity.
Keywords: doxorubicin; human organoid models; molecular mechanisms; toxicity; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

