The Transmembrane Protease TMPRSS2 as a Therapeutic Target for COVID-19 Treatment
- PMID: 35163273
- PMCID: PMC8836196
- DOI: 10.3390/ijms23031351
The Transmembrane Protease TMPRSS2 as a Therapeutic Target for COVID-19 Treatment
Abstract
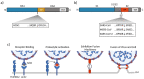
TMPRSS2 is a type II transmembrane protease with broad expression in epithelial cells of the respiratory and gastrointestinal tract, the prostate, and other organs. Although the physiological role of TMPRSS2 remains largely elusive, several endogenous substrates have been identified. TMPRSS2 serves as a major cofactor in SARS-CoV-2 entry, and primes glycoproteins of other respiratory viruses as well. Consequently, inhibiting TMPRSS2 activity is a promising strategy to block viral infection. In this review, we provide an overview of the role of TMPRSS2 in the entry processes of different respiratory viruses. We then review the different classes of TMPRSS2 inhibitors and their clinical development, with a focus on COVID-19 treatment.
Keywords: SARS-CoV-2; TMPRSS2; coronavirus; influenza; protease inhibitor.
Conflict of interest statement
L.W. and J.M. are inventors on patent applications claiming to use TMPRSS2-inhibitory peptidomimetics, peptides and proteins as antivirals.
Figures



References
-
- Barré O., Dufour A., Eckhard U., Kappelhoff R., Béliveau F., Leduc R., Overall C.M. Cleavage specificity analysis of six type II transmembrane serine proteases (TTSPs) using PICS with proteome-derived peptide libraries. PLoS ONE. 2014;9:e105984. doi: 10.1371/journal.pone.0105984. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

