Anti-Excitotoxic Effects of N-Butylidenephthalide Revealed by Chemically Insulted Purkinje Progenitor Cells Derived from SCA3 iPSCs
- PMID: 35163312
- PMCID: PMC8836169
- DOI: 10.3390/ijms23031391
Anti-Excitotoxic Effects of N-Butylidenephthalide Revealed by Chemically Insulted Purkinje Progenitor Cells Derived from SCA3 iPSCs
Abstract
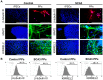
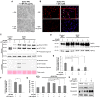
Spinocerebellar ataxia type 3 (SCA3) is characterized by the over-repetitive CAG codon in the ataxin-3 gene (ATXN3), which encodes the mutant ATXN3 protein. The pathological defects of SCA3 such as the impaired aggresomes, autophagy, and the proteasome have been reported previously. To date, no effective treatment is available for SCA3 disease. This study aimed to study anti-excitotoxic effects of n-butylidenephthalide by chemically insulted Purkinje progenitor cells derived from SCA3 iPSCs. We successfully generated Purkinje progenitor cells (PPs) from SCA3 patient-derived iPSCs. The PPs, expressing both neural and Purkinje progenitor's markers, were acquired after 35 days of differentiation. In comparison with the PPs derived from control iPSCs, SCA3 iPSCs-derived PPs were more sensitive to the excitotoxicity induced by quinolinic acid (QA). The observations of QA-treated SCA3 PPs showing neural degeneration including neurite shrinkage and cell number decrease could be used to quickly and efficiently identify drug candidates. Given that the QA-induced neural cell death of SCA3 PPs was established, the activity of calpain in SCA3 PPs was revealed. Furthermore, the expression of cleaved poly (ADP-ribose) polymerase 1 (PARP1), a marker of apoptotic pathway, and the accumulation of ATXN3 proteolytic fragments were observed. When SCA3 PPs were treated with n-butylidenephthalide (n-BP), upregulated expression of calpain 2 and concurrent decreased level of calpastatin could be reversed, and the overall calpain activity was accordingly suppressed. Such findings reveal that n-BP could not only inhibit the cleavage of ATXN3 but also protect the QA-induced excitotoxicity from the Purkinje progenitor loss.
Keywords: ATXN3; Purkinje progenitor; SCA3; iPSCs; n-butylidenephthalide; quinolinic acid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
n-Butylidenephthalide exhibits protection against neurotoxicity through regulation of tryptophan 2, 3 dioxygenase in spinocerebellar ataxia type 3.Neuropharmacology. 2017 May 1;117:434-446. doi: 10.1016/j.neuropharm.2017.02.014. Epub 2017 Feb 20. Neuropharmacology. 2017. PMID: 28223212
-
n-Butylidenephthalide Modulates Autophagy to Ameliorate Neuropathological Progress of Spinocerebellar Ataxia Type 3 through mTOR Pathway.Int J Mol Sci. 2021 Jun 13;22(12):6339. doi: 10.3390/ijms22126339. Int J Mol Sci. 2021. PMID: 34199295 Free PMC article.
-
Autophagy Promoted the Degradation of Mutant ATXN3 in Neurally Differentiated Spinocerebellar Ataxia-3 Human Induced Pluripotent Stem Cells.Biomed Res Int. 2016;2016:6701793. doi: 10.1155/2016/6701793. Epub 2016 Oct 25. Biomed Res Int. 2016. PMID: 27847820 Free PMC article.
-
Toward therapeutic targets for SCA3: Insight into the role of Machado-Joseph disease protein ataxin-3 in misfolded proteins clearance.Prog Neurobiol. 2015 Sep;132:34-58. doi: 10.1016/j.pneurobio.2015.06.004. Epub 2015 Jun 27. Prog Neurobiol. 2015. PMID: 26123252 Review.
-
Planning Future Clinical Trials for Machado-Joseph Disease.Adv Exp Med Biol. 2018;1049:321-348. doi: 10.1007/978-3-319-71779-1_17. Adv Exp Med Biol. 2018. PMID: 29427112 Review.
Cited by
-
n-Butylidenephthalide recovered calcium homeostasis to ameliorate neurodegeneration of motor neurons derived from amyotrophic lateral sclerosis iPSCs.PLoS One. 2024 Nov 7;19(11):e0311573. doi: 10.1371/journal.pone.0311573. eCollection 2024. PLoS One. 2024. PMID: 39509425 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

