Fine Mapping and Candidate Gene Prediction of Tuber Shape Controlling Ro Locus Based on Integrating Genetic and Transcriptomic Analyses in Potato
- PMID: 35163389
- PMCID: PMC8836246
- DOI: 10.3390/ijms23031470
Fine Mapping and Candidate Gene Prediction of Tuber Shape Controlling Ro Locus Based on Integrating Genetic and Transcriptomic Analyses in Potato
Abstract
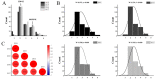
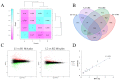
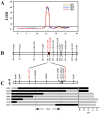
Tuber shape is one of the most important quality traits in potato appearance. Since poor or irregular shape results in higher costs for processing and influences the consumers' willingness to purchase, breeding for shape uniformity and shallow eye depth is highly important. Previous studies showed that the major round tuber shape controlling locus, the Ro locus, is located on chromosome 10. However, fine mapping and cloning of tuber shape genes have not been reported. In this study, the analyses of tissue sectioning and transcriptome sequencing showed that the developmental differences between round and elongated tuber shapes begin as early as the hook stage of the stolon. To fine map tuber shape genes, a high-density genetic linkage map of the Ro region on chromosome 10 based on a diploid segregating population was constructed. The total length of the genetic linkage map was 25.8 cM and the average marker interval was 1.98 cM. Combined with phenotypic data collected from 2014 to 2017, one major quantitative trait locus (QTL) for tuber shape was identified, which explained 61.7-72.9% of the tuber shape variation. Through the results of genotyping and phenotypic investigation of recombinant individuals, Ro was fine mapped in a 193.43 kb interval, which contained 18 genes. Five candidate genes were preliminarily predicted based on tissue sections and transcriptome sequencing. This study provides an important basis for cloning Ro gene(s).
Keywords: QTL; Solanum tuberosum L.; candidate gene prediction; tuber shape.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Hirsch C.N., Hirsch C.D., Felcher K., Coombs J., Zarka D., Deynze A.V., Jong W.D., Veilleux R.E., Jansky S., Bethke P. Retrospective View of North American Potato (Solanum tuberosum L.) Breeding in the 20th and 21st Centuries. G3 Genes Genomes Genet. 2013;3:1003–1013. doi: 10.1534/g3.113.005595. - DOI - PMC - PubMed
-
- Si Y., Sankaran S., Knowles N.R., Pavek M.J. Potato Tuber Length-Width Ratio Assessment Using Image Analysis. Am. J. Potato Res. 2016;94:88–93. doi: 10.1007/s12230-016-9545-1. - DOI
-
- Jong H.D., Burns V.J. Inheritance of tuber shape in cultivated diploid potatoes. Am. Potato J. 1993;70:267–284. doi: 10.1007/BF02849314. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

