Allosterism in the PDZ Family
- PMID: 35163402
- PMCID: PMC8836106
- DOI: 10.3390/ijms23031454
Allosterism in the PDZ Family
Abstract
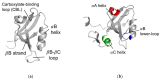
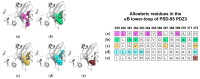
Dynamic allosterism allows the propagation of signal throughout a protein. The PDZ (PSD-95/Dlg1/ZO-1) family has been named as a classic example of dynamic allostery in small modular domains. While the PDZ family consists of more than 200 domains, previous efforts have primarily focused on a few well-studied PDZ domains, including PTP-BL PDZ2, PSD-95 PDZ3, and Par6 PDZ. Taken together, experimental and computational studies have identified regions of these domains that are dynamically coupled to ligand binding. These regions include the αA helix, the αB lower-loop, and the αC helix. In this review, we summarize the specific residues on the αA helix, the αB lower-loop, and the αC helix of PTP-BL PDZ2, PSD-95 PDZ3, and Par6 PDZ that have been identified as participants in dynamic allostery by either experimental or computational approaches. This review can serve as an index for researchers to look back on the previously identified allostery in the PDZ family. Interestingly, our summary of previous work reveals clear consistencies between the domains. While the PDZ family has a low sequence identity, we show that some of the most consistently identified allosteric residues within PTP-BL PDZ2 and PSD-95 PDZ3 domains are evolutionarily conserved. These residues include A46/A347, V61/V362, and L66/L367 on PTP-BL PDZ2 and PSD-95 PDZ3, respectively. Finally, we expose a need for future work to explore dynamic allostery within (1) PDZ domains with multiple binding partners and (2) multidomain constructs containing a PDZ domain.
Keywords: PDZ domain; allosterism; dynamic allostery; key residues.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




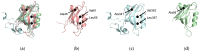
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

