Regulation of the Emissions of the Greenhouse Gas Nitrous Oxide by the Soybean Endosymbiont Bradyrhizobium diazoefficiens
- PMID: 35163408
- PMCID: PMC8836242
- DOI: 10.3390/ijms23031486
Regulation of the Emissions of the Greenhouse Gas Nitrous Oxide by the Soybean Endosymbiont Bradyrhizobium diazoefficiens
Abstract
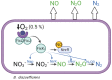
The greenhouse gas nitrous oxide (N2O) has strong potential to drive climate change. Soils are a major source of N2O, with microbial nitrification and denitrification being the primary processes involved in such emissions. The soybean endosymbiont Bradyrhizobium diazoefficiens is a model microorganism to study denitrification, a process that depends on a set of reductases, encoded by the napEDABC, nirK, norCBQD, and nosRZDYFLX genes, which sequentially reduce nitrate (NO3-) to nitrite (NO2-), nitric oxide (NO), N2O, and dinitrogen (N2). In this bacterium, the regulatory network and environmental cues governing the expression of denitrification genes rely on the FixK2 and NnrR transcriptional regulators. To understand the role of FixK2 and NnrR proteins in N2O turnover, we monitored real-time kinetics of NO3-, NO2-, NO, N2O, N2, and oxygen (O2) in a fixK2 and nnrR mutant using a robotized incubation system. We confirmed that FixK2 and NnrR are regulatory determinants essential for NO3- respiration and N2O reduction. Furthermore, we demonstrated that N2O reduction by B. diazoefficiens is independent of canonical inducers of denitrification, such as the nitrogen oxide NO3-, and it is negatively affected by acidic and alkaline conditions. These findings advance the understanding of how specific environmental conditions and two single regulators modulate N2O turnover in B. diazoefficiens.
Keywords: denitrification; dinitrogen; gene expression; nitric oxide; nitrous oxide reductase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Van Spanning R.J.M., Richardson D.J., Ferguson S.J. Introduction to the biochemistry and molecular biology of denitrification. In: Bothe H., Ferguson S.J., Newton W.E., editors. Biology of the Nitrogen Cycle. 1st ed. Elsevier; Amsterdam, The Netherlands: 2007. pp. 3–20.
-
- Conrad R. Metabolism of Nitric Oxide in Soil and Soil Microorganisms and Regulation of Flux into the Atmosphere. Microbiol. Atmos. Trace Gases. 1996;60:167–203. doi: 10.1007/978-3-642-61096-7_11. - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

