Metallothionein1A Regulates Rhizobial Infection and Nodulation in Phaseolus vulgaris
- PMID: 35163415
- PMCID: PMC8836284
- DOI: 10.3390/ijms23031491
Metallothionein1A Regulates Rhizobial Infection and Nodulation in Phaseolus vulgaris
Abstract
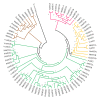
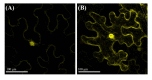
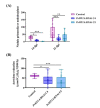
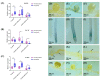
Metallothioneins (MTs) constitute a heterogeneous family of ubiquitous metal ion-binding proteins. In plants, MTs participate in the regulation of cell growth and proliferation, protection against heavy metal stress, oxidative stress responses, and responses to pathogen attack. Despite their wide variety of functions, the role of MTs in symbiotic associations, specifically nodule-fabacean symbiosis, is poorly understood. Here, we analyzed the role of the PvMT1A gene in Phaseolus vulgaris-Rhizobium tropici symbiosis using bioinformatics and reverse genetics approaches. Using in silico analysis, we identified six genes encoding MTs in P. vulgaris, which were clustered into three of the four classes described in plants. PvMT1A transcript levels were significantly higher in roots inoculated with R. tropici at 7 and 30 days post inoculation (dpi) than in non-inoculated roots. Functional analysis showed that downregulating PvMT1A by RNA interference (RNAi) reduced the number of infection events at 7 and 10 dpi and the number of nodules at 14 and 21 dpi. In addition, nodule development was negatively affected in PvMT1A:RNAi transgenic roots, and these nodules displayed a reduced nitrogen fixation rate at 21 dpi. These results strongly suggest that PvMT1A plays an important role in the infection process and nodule development in P. vulgaris during rhizobial symbiosis.
Keywords: common bean; metallothionein; nodule symbiosis; reactive oxygen species; rhizobial infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

