Induction of PLXNA4 Gene during Neural Differentiation in Human Umbilical-Cord-Derived Mesenchymal Stem Cells by Low-Intensity Sub-Sonic Vibration
- PMID: 35163445
- PMCID: PMC8835879
- DOI: 10.3390/ijms23031522
Induction of PLXNA4 Gene during Neural Differentiation in Human Umbilical-Cord-Derived Mesenchymal Stem Cells by Low-Intensity Sub-Sonic Vibration
Abstract
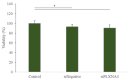
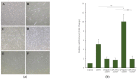
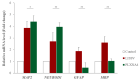
Human umbilical-cord-derived mesenchymal stem cells (hUC-MSC) are a type of mesenchymal stem cells and are more primitive than other MSCs. In this study, we identify novel genes and signal-activating proteins involved in the neural differentiation of hUC-MSCs induced by Low-Intensity Sub-Sonic Vibration (LISSV). RNA sequencing was used to find genes involved in the differentiation process by LISSV. The changes in hUC-MSCs caused by LISSV were confirmed by PLXNA4 overexpression and gene knockdown through small interfering RNA experiments. The six genes were increased among genes related to neurons and the nervous system. One of them, the PLXNA4 gene, is known to play a role as a guide for axons in the development of the nervous system. When the PLXNA4 recombinant protein was added, neuron-related genes were increased. In the PLXNA4 gene knockdown experiment, the expression of neuron-related genes was not changed by LISSV exposure. The PLXNA4 gene is activated by sema family ligands. The expression of SEMA3A was increased by LISSV, and its downstream signaling molecule, FYN, was also activated. We suggest that the PLXNA4 gene plays an important role in hUC-MSC neuronal differentiation through exposure to LISSV. The differentiation process depends on SEMA3A-PLXNA4-dependent FYN activation in hUC-MSCs.
Keywords: PLXNA4; SEMA3A; hUC-MSCs; low-intensity sub-sonic vibration; neural signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






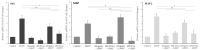




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

