Whole-Genome Sequencing and RNA-Seq Reveal Differences in Genetic Mechanism for Flowering Response between Weedy Rice and Cultivated Rice
- PMID: 35163531
- PMCID: PMC8836195
- DOI: 10.3390/ijms23031608
Whole-Genome Sequencing and RNA-Seq Reveal Differences in Genetic Mechanism for Flowering Response between Weedy Rice and Cultivated Rice
Abstract
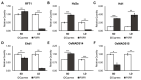
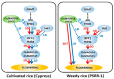
Flowering is a key agronomic trait that influences adaptation and productivity. Previous studies have indicated the genetic complexity associated with the flowering response in a photoinsensitive weedy rice accession PSRR-1 despite the presence of a photosensitive allele of a key flowering gene Hd1. In this study, we used whole-genome and RNA sequencing data from both cultivated and weedy rice to add further insights. The de novo assembly of unaligned sequences predicted 225 genes, in which 45 were specific to PSRR-1, including two genes associated with flowering. Comparison of the variants in PSRR-1 with the 3K rice genome (RG) dataset identified unique variants within the heading date QTLs. Analyses of the RNA-Seq result under both short-day (SD) and long-day (LD) conditions revealed that many differentially expressed genes (DEGs) colocalized with the flowering QTLs, and some DEGs such as Hd1, OsMADS56, Hd3a, and RFT1 had unique variants in PSRR-1. Ehd1, Hd1, OsMADS15, and OsMADS56 showed different alternate splicing (AS) events between genotypes and day length conditions. OsMADS56 was expressed in PSRR-1 but not in Cypress under both LD and SD conditions. Based on variations in both sequence and expression, the unique flowering response in PSRR-1 may be due to the high-impact variants of flowering genes, and OsMADS56 is proposed as a key regulator for its day-neutral flowering response.
Keywords: Oryza sativa; RNA-Sequencing; days to heading; genetic interaction; photosensitivity; red rice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
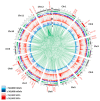
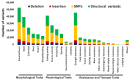
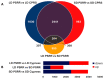

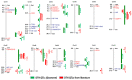

References
-
- Delouche J.C., Labrada R., Burgos N.R., Gealy D.R. FAO Plant Production and Protection Paper 188. FAO; Rome, Italy: 2007. Weedy rices: Origin, biology, ecology and control.144p
-
- Vaughan D.A., Lu B.R., Tomooka N. Was Asian rice (Oryza sativa) domesticated more than once? Rice. 2008;1:16–24. doi: 10.1007/s12284-008-9000-0. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

