Genome-Wide Analysis Indicates a Complete Prostaglandin Pathway from Synthesis to Inactivation in Pacific White Shrimp, Litopenaeus vannamei
- PMID: 35163575
- PMCID: PMC8835781
- DOI: 10.3390/ijms23031654
Genome-Wide Analysis Indicates a Complete Prostaglandin Pathway from Synthesis to Inactivation in Pacific White Shrimp, Litopenaeus vannamei
Abstract
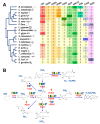
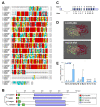
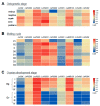
Prostaglandins (PGs) play many essential roles in the development, immunity, metabolism, and reproduction of animals. In vertebrates, arachidonic acid (ARA) is generally converted to prostaglandin G2 (PGG2) and H2 (PGH2) by cyclooxygenase (COX); then, various biologically active PGs are produced through different downstream prostaglandin synthases (PGSs), while PGs are inactivated by 15-hydroxyprostaglandin dehydrogenase (PGDH). However, there is very limited knowledge of the PG biochemical pathways in invertebrates, particularly for crustaceans. In this study, nine genes involved in the prostaglandin pathway, including a COX, seven PGSs (PGES, PGES2, PGDS1/2, PGFS, AKR1C3, and TXA2S), and a PGDH were identified based on the Pacific white shrimp (Litopenaeus vannamei) genome, indicating a more complete PG pathway from synthesis to inactivation in crustaceans than in insects and mollusks. The homologous genes are conserved in amino acid sequences and structural domains, similar to those of related species. The expression patterns of these genes were further analyzed in a variety of tissues and developmental processes by RNA sequencing and quantitative real-time PCR. The mRNA expression of PGES was relatively stable in various tissues, while other genes were specifically expressed in distant tissues. During embryo development to post-larvae, COX, PGDS1, GDS2, and AKR1C3 expressions increased significantly, and increasing trends were also observed on PGES, PGDS2, and AKR1C3 at the post-molting stage. During the ovarian maturation, decreasing trends were found on PGES1, PGDS2, and PGDH in the hepatopancreas, but all gene expressions remained relatively stable in ovaries. In conclusion, this study provides basic knowledge for the synthesis and inactivation pathway of PG in crustaceans, which may contribute to the understanding of their regulatory mechanism in ontogenetic development and reproduction.
Keywords: crustacean; genome-wide analysis; mRNA expression; prostaglandin; synthesis and inactivation.
Conflict of interest statement
Q.G. from EasyATGC Limited Liability Company download the genomic data from Genbank and annotated the genes for the study. EasyATGC Limited Liability Company had no role in the topic selected, experimental design, execution, interpretation, or writing of the study. No fund of the study was supported by EasyATGC Limited Liability Company, and the study did not refer to any product from EasyATGC Limited Liability Company.
Figures


References
MeSH terms
Substances
Grants and funding
- 41876196/National Natural Science Foundation of China
- 32172954/National Natural Science Foundation of China
- 201904020009/Guangzhou Municipal Science and Technology Bureau
- 2018YFD0900100/Ministry of science and technology of China
- 2019KJ149/Department of Agriculture and Rural Affairs of Guangdong Province
LinkOut - more resources
Full Text Sources
Miscellaneous

