Model of Genetic Code Structure Evolution under Various Types of Codon Reading
- PMID: 35163612
- PMCID: PMC8835785
- DOI: 10.3390/ijms23031690
Model of Genetic Code Structure Evolution under Various Types of Codon Reading
Abstract
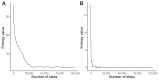
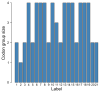
The standard genetic code (SGC) is a set of rules according to which 64 codons are assigned to 20 canonical amino acids and stop coding signal. As a consequence, the SGC is redundant because there is a greater number of codons than the number of encoded labels. This redundancy implies the existence of codons that encode the same genetic information. The size and organization of such synonymous codon blocks are important characteristics of the SGC structure whose evolution is still unclear. Therefore, we studied possible evolutionary mechanisms of the codon block structure. We conducted computer simulations assuming that coding systems at early stages of the SGC evolution were sets of ambiguous codon assignments with high entropy. We included three types of reading systems characterized by different inaccuracy and pattern of codon recognition. In contrast to the previous study, we allowed for evolution of the reading systems and their competition. The simulations performed under minimization of translational errors and reduction of coding ambiguity produced the coding system resistant to these errors. The reading system similar to that present in the SGC dominated the others very quickly. The survived system was also characterized by low entropy and possessed properties similar to that in the SGC. Our simulation show that the unambiguous SGC could emerged from a code with a lower level of ambiguity and the number of tRNAs increased during the evolution.
Keywords: amino acid; codon; evolution; genetic code.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
The influence of different types of translational inaccuracies on the genetic code structure.BMC Bioinformatics. 2019 Mar 6;20(1):114. doi: 10.1186/s12859-019-2661-4. BMC Bioinformatics. 2019. PMID: 30841864 Free PMC article.
-
Evolution of the genetic code through progressive symmetry breaking.J Theor Biol. 2014 Apr 21;347:95-108. doi: 10.1016/j.jtbi.2014.01.002. Epub 2014 Jan 14. J Theor Biol. 2014. PMID: 24434741
-
Models of genetic code structure evolution with variable number of coded labels.Biosystems. 2021 Dec;210:104528. doi: 10.1016/j.biosystems.2021.104528. Epub 2021 Sep 4. Biosystems. 2021. PMID: 34492316
-
Origin and Evolution of the Universal Genetic Code.Annu Rev Genet. 2017 Nov 27;51:45-62. doi: 10.1146/annurev-genet-120116-024713. Epub 2017 Aug 30. Annu Rev Genet. 2017. PMID: 28853922 Review.
-
Origin and evolution of the genetic code: the universal enigma.IUBMB Life. 2009 Feb;61(2):99-111. doi: 10.1002/iub.146. IUBMB Life. 2009. PMID: 19117371 Free PMC article. Review.
Cited by
-
The Origin of Translation: Bridging the Nucleotides and Peptides.Int J Mol Sci. 2022 Dec 22;24(1):197. doi: 10.3390/ijms24010197. Int J Mol Sci. 2022. PMID: 36613641 Free PMC article. Review.
-
The Role of Mutations, Addition of Amino Acids, and Exchange of Genetic Information in the Coevolution of Primitive Coding Systems.Int J Mol Sci. 2025 Jul 25;26(15):7176. doi: 10.3390/ijms26157176. Int J Mol Sci. 2025. PMID: 40806309 Free PMC article.
References
-
- Barbieri M. Evolution of the Genetic Code: The Ribosome-Oriented Model. Biol. Theory. 2015;10:301–310. doi: 10.1007/s13752-015-0225-z. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

