A Multi-Disulfide Receptor-Binding Domain (RBD) of the SARS-CoV-2 Spike Protein Expressed in E. coli Using a SEP-Tag Produces Antisera Interacting with the Mammalian Cell Expressed Spike (S1) Protein
- PMID: 35163624
- PMCID: PMC8835783
- DOI: 10.3390/ijms23031703
A Multi-Disulfide Receptor-Binding Domain (RBD) of the SARS-CoV-2 Spike Protein Expressed in E. coli Using a SEP-Tag Produces Antisera Interacting with the Mammalian Cell Expressed Spike (S1) Protein
Abstract
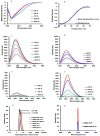
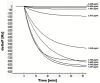
An Escherichia coli (E. coli) production of the receptor-binding domain (RBD) of the SARS-CoV-2 (isolate Wuhan-Hu-1) spike protein would significantly accelerate the search for anti-COVID-19 therapeutics because of its versatility and low cost. However, RBD contains four disulfide bonds and its expression in E. coli is limited by the formation of aberrant disulfide bonds resulting in inclusion bodies. Here, we show that a solubility-enhancing peptide (SEP) tag containing nine arginine residues (RBD-C9R) attached at the C-terminus can overcome this problem. The SEP-tag increased the expression in the soluble fraction and the final yield by five times (2 mg/L). The folding properties of the E. coli expressed RBD-C9R were demonstrated with biophysical characterization using RP-HPLC, circular dichroism, thermal denaturation, fluorescence, and light scattering. A quartz crystal microbalance (QCM) analysis confirmed the binding activity of RBD-C9R with ACE2, the host cell's receptor. In addition, RBD-C9R elicited a Th-2 immune response with a high IgG titer in Jcl: ICR mice. The RBD-C9R antisera interacted with both itself and the mammalian-cell expressed spike protein (S1), as demonstrated by ELISA, indicating that the E. coli expressed RBD-C9R harbors native-like epitopes. Overall, these results emphasize the potential of our SEP-tag for the E. coli production of active multi-disulfide-bonded RBD.
Keywords: Escherichia coli expression; disulfide bond; fusion tag; immunogenicity; solubility.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
EGFR extracellular domain III expressed in Escherichia coli with SEP tag shows improved biophysical and functional properties and generate anti-sera inhibiting cancer cell growth.Biochem Biophys Res Commun. 2021 May 28;555:121-127. doi: 10.1016/j.bbrc.2021.03.102. Epub 2021 Apr 1. Biochem Biophys Res Commun. 2021. PMID: 33813270
-
An Escherichia coli Expressed Multi-Disulfide Bonded SARS-CoV-2 RBD Shows Native-like Biophysical Properties and Elicits Neutralizing Antisera in a Mouse Model.Int J Mol Sci. 2022 Dec 12;23(24):15744. doi: 10.3390/ijms232415744. Int J Mol Sci. 2022. PMID: 36555383 Free PMC article.
-
Non-Glycosylated SARS-CoV-2 Omicron BA.5 Receptor Binding Domain (RBD) with a Native-like Conformation Induces a Robust Immune Response with Potent Neutralization in a Mouse Model.Molecules. 2024 Jun 5;29(11):2676. doi: 10.3390/molecules29112676. Molecules. 2024. PMID: 38893549 Free PMC article.
-
Antisera Produced Using an E. coli-Expressed SARS-CoV-2 RBD and Complemented with a Minimal Dose of Mammalian-Cell-Expressed S1 Subunit of the Spike Protein Exhibits Improved Neutralization.Int J Mol Sci. 2023 Jun 24;24(13):10583. doi: 10.3390/ijms241310583. Int J Mol Sci. 2023. PMID: 37445760 Free PMC article.
-
Disulfide Bonds Play a Critical Role in the Structure and Function of the Receptor-binding Domain of the SARS-CoV-2 Spike Antigen.J Mol Biol. 2022 Jan 30;434(2):167357. doi: 10.1016/j.jmb.2021.167357. Epub 2021 Nov 12. J Mol Biol. 2022. PMID: 34780781 Free PMC article.
Cited by
-
Amyloidogenesis of SARS-CoV-2 delta plus and omicron variants receptor-binding domain (RBD): impact of SUMO fusion tag.Biotechnol Lett. 2024 Dec;46(6):1037-1048. doi: 10.1007/s10529-024-03525-9. Epub 2024 Aug 25. Biotechnol Lett. 2024. PMID: 39182215
-
The Immunogenicity of DENV1-4 ED3s Strongly Differ despite Their Almost Identical Three-Dimensional Structures and High Sequence Similarities.Int J Mol Sci. 2023 Jan 25;24(3):2393. doi: 10.3390/ijms24032393. Int J Mol Sci. 2023. PMID: 36768719 Free PMC article.
-
A Live-Cell Imaging-Based Fluorescent SARS-CoV-2 Neutralization Assay by Antibody-Mediated Blockage of Receptor Binding Domain-ACE2 Interaction.BioTech (Basel). 2025 Feb 14;14(1):10. doi: 10.3390/biotech14010010. BioTech (Basel). 2025. PMID: 39982277 Free PMC article.
-
SARS-CoV-2 spike trimer vaccine expressed in Nicotiana benthamiana adjuvanted with Alum elicits protective immune responses in mice.Plant Biotechnol J. 2022 Dec;20(12):2298-2312. doi: 10.1111/pbi.13908. Epub 2022 Sep 5. Plant Biotechnol J. 2022. PMID: 36062974 Free PMC article.
-
Protein Expression Platforms and the Challenges of Viral Antigen Production.Vaccines (Basel). 2024 Nov 28;12(12):1344. doi: 10.3390/vaccines12121344. Vaccines (Basel). 2024. PMID: 39772006 Free PMC article. Review.
References
-
- Salzer R., Clark J.J., Vaysburd M., Chang V.T., Albecka A., Kiss L., Sharma P., Gonzalez Llamazares A., Kipar A., Hiscox J.A., et al. Single-dose immunisation with a multimerised SARS-CoV-2 receptor binding domain (RBD) induces an enhanced and protective response in mice. FEBS Lett. 2021;595:2323–2340. doi: 10.1002/1873-3468.14171. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

