Genome-Wide Prediction of Transcription Start Sites in Conifers
- PMID: 35163661
- PMCID: PMC8836283
- DOI: 10.3390/ijms23031735
Genome-Wide Prediction of Transcription Start Sites in Conifers
Abstract
The identification of promoters is an essential step in the genome annotation process, providing a framework for gene regulatory networks and their role in transcription regulation. Despite considerable advances in the high-throughput determination of transcription start sites (TSSs) and transcription factor binding sites (TFBSs), experimental methods are still time-consuming and expensive. Instead, several computational approaches have been developed to provide fast and reliable means for predicting the location of TSSs and regulatory motifs on a genome-wide scale. Numerous studies have been carried out on the regulatory elements of mammalian genomes, but plant promoters, especially in gymnosperms, have been left out of the limelight and, therefore, have been poorly investigated. The aim of this study was to enhance and expand the existing genome annotations using computational approaches for genome-wide prediction of TSSs in the four conifer species: loblolly pine, white spruce, Norway spruce, and Siberian larch. Our pipeline will be useful for TSS predictions in other genomes, especially for draft assemblies, where reliable TSS predictions are not usually available. We also explored some of the features of the nucleotide composition of the predicted promoters and compared the GC properties of conifer genes with model monocot and dicot plants. Here, we demonstrate that even incomplete genome assemblies and partial annotations can be a reliable starting point for TSS annotation. The results of the TSS prediction in four conifer species have been deposited in the Persephone genome browser, which allows smooth visualization and is optimized for large data sets. This work provides the initial basis for future experimental validation and the study of the regulatory regions to understand gene regulation in gymnosperms.
Keywords: TATA-box; conifer; gymnosperms; promoter prediction; transcription factor binding site; transcription start site.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

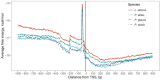





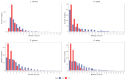
Similar articles
-
GPMiner: an integrated system for mining combinatorial cis-regulatory elements in mammalian gene group.BMC Genomics. 2012;13 Suppl 1(Suppl 1):S3. doi: 10.1186/1471-2164-13-S1-S3. Epub 2012 Jan 17. BMC Genomics. 2012. PMID: 22369687 Free PMC article.
-
Paired-end analysis of transcription start sites in Arabidopsis reveals plant-specific promoter signatures.Plant Cell. 2014 Jul;26(7):2746-60. doi: 10.1105/tpc.114.125617. Epub 2014 Jul 17. Plant Cell. 2014. PMID: 25035402 Free PMC article.
-
Genome-wide computational prediction and analysis of core promoter elements across plant monocots and dicots.PLoS One. 2013 Oct 29;8(10):e79011. doi: 10.1371/journal.pone.0079011. eCollection 2013. PLoS One. 2013. PMID: 24205361 Free PMC article.
-
Computational annotation of miRNA transcription start sites.Brief Bioinform. 2021 Jan 18;22(1):380-392. doi: 10.1093/bib/bbz178. Brief Bioinform. 2021. PMID: 32003428 Free PMC article. Review.
-
Role of DNA sequence based structural features of promoters in transcription initiation and gene expression.Curr Opin Struct Biol. 2014 Apr;25:77-85. doi: 10.1016/j.sbi.2014.01.007. Epub 2014 Feb 4. Curr Opin Struct Biol. 2014. PMID: 24503515 Review.
Cited by
-
The Complete Chloroplast Genome Sequence of Laportea bulbifera (Sieb. et Zucc.) Wedd. and Comparative Analysis with Its Congeneric Species.Genes (Basel). 2022 Nov 28;13(12):2230. doi: 10.3390/genes13122230. Genes (Basel). 2022. PMID: 36553498 Free PMC article.
-
Classification of Promoter Sequences from Human Genome.Int J Mol Sci. 2023 Aug 8;24(16):12561. doi: 10.3390/ijms241612561. Int J Mol Sci. 2023. PMID: 37628742 Free PMC article.
-
Plant Biology and Biotechnology: Focus on Genomics and Bioinformatics.Int J Mol Sci. 2022 Jun 17;23(12):6759. doi: 10.3390/ijms23126759. Int J Mol Sci. 2022. PMID: 35743200 Free PMC article.
-
Database of Potential Promoter Sequences in the Capsicum annuum Genome.Biology (Basel). 2022 Jul 26;11(8):1117. doi: 10.3390/biology11081117. Biology (Basel). 2022. PMID: 35892972 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

