Clinical Utility of a Unique Genome-Wide DNA Methylation Signature for KMT2A-Related Syndrome
- PMID: 35163737
- PMCID: PMC8836705
- DOI: 10.3390/ijms23031815
Clinical Utility of a Unique Genome-Wide DNA Methylation Signature for KMT2A-Related Syndrome
Abstract
Wiedemann-Steiner syndrome (WDSTS) is a Mendelian syndromic intellectual disability (ID) condition associated with hypertrichosis cubiti, short stature, and characteristic facies caused by pathogenic variants in the KMT2A gene. Clinical features can be inconclusive in mild and unusual WDSTS presentations with variable ID (mild to severe), facies (typical or not) and other associated malformations (bone, cerebral, renal, cardiac and ophthalmological anomalies). Interpretation and classification of rare KMT2A variants can be challenging. A genome-wide DNA methylation episignature for KMT2A-related syndrome could allow functional classification of variants and provide insights into the pathophysiology of WDSTS. Therefore, we assessed genome-wide DNA methylation profiles in a cohort of 60 patients with clinical diagnosis for WDSTS or Kabuki and identified a unique highly sensitive and specific DNA methylation episignature as a molecular biomarker of WDSTS. WDSTS episignature enabled classification of variants of uncertain significance in the KMT2A gene as well as confirmation of diagnosis in patients with clinical presentation of WDSTS without known genetic variants. The changes in the methylation profile resulting from KMT2A mutations involve global reduction in methylation in various genes, including homeobox gene promoters. These findings provide novel insights into the molecular etiology of WDSTS and explain the broad phenotypic spectrum of the disease.
Keywords: DNA methylation; KMT2A gene; Wiedemann–Steiner syndrome; epigenetics; episignature; intellectual disability; neurodevelopmental disorders.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
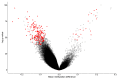

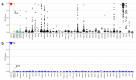

References
-
- Lebrun N., Giurgea I., Goldenberg A., Dieux A., Afenjar A., Ghoumid J., Diebold B., Mietton L., Briand-Suleau A., Billuart P., et al. Molecular and cellular issues of KMT2A variants involved in Wiedemann-Steiner syndrome. Eur. J. Hum. Genet. 2017;26:107–116. doi: 10.1038/s41431-017-0033-y. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- Beyond Genomics: Assessing the Improvement in Diagnosis of Rare Diseases using Clinical Epigenomics in Canada, EpiSign-CAN/London Health Sciences Molecular Diagnostics Development Fund and Genome Canada Genomic Applications Partnership Program Grant
- CCR-2017-23669081/Italian Ministry of Health
- RCR-2020-23670068_001/Italian Ministry of Health
- Ricerca Corrente 2021/Italian Ministry of Health
- FOE 2019/Italian Ministry of Research
LinkOut - more resources
Full Text Sources
Research Materials

