Structural and Functional Differences between Homologous Bacterial Ribonucleases
- PMID: 35163789
- PMCID: PMC8837141
- DOI: 10.3390/ijms23031867
Structural and Functional Differences between Homologous Bacterial Ribonucleases
Abstract
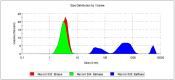
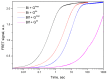
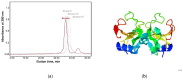
Small cationic guanyl-preferring ribonucleases (RNases) produced by the Bacillus species share a similar protein tertiary structure with a high degree of amino acid sequence conservation. However, they form dimers that differ in conformation and stability. Here, we have addressed the issues (1) whether the homologous RNases also have distinctions in catalytic activity towards different RNA substrates and interactions with the inhibitor protein barstar, and (2) whether these differences correlate with structural features of the proteins. Circular dichroism and dynamic light scattering assays revealed distinctions in the structures of homologous RNases. The activity levels of the RNases towards natural RNA substrates, as measured spectrometrically by acid-soluble hydrolysis products, were similar and decreased in the row high-polymeric RNA >>> transport RNA > double-stranded RNA. However, stopped flow kinetic studies on model RNA substrates containing the guanosine residue in a hairpin stem or a loop showed that the cleavage rates of these enzymes were different. Moreover, homologous RNases were inhibited by the barstar with diverse efficiency. Therefore, minor changes in structure elements of homologous proteins have a potential to significantly effect molecule stability and functional activities, such as catalysis or ligand binding.
Keywords: balifase; balnase; barnase; barstar; binase; catalytic activity; ribonuclease; ribonuclease inhibitor; structural organization.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



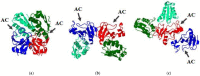
References
-
- Takahashi K., Moore S. 13 Ribonuclease T1. Enzymes. 1982;15:435–467.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

