The Competitive Endogenous RNA (ceRNA) Regulation in Porcine Alveolar Macrophages (3D4/21) Infected by Swine Influenza Virus (H1N1 and H3N2)
- PMID: 35163797
- PMCID: PMC8836399
- DOI: 10.3390/ijms23031875
The Competitive Endogenous RNA (ceRNA) Regulation in Porcine Alveolar Macrophages (3D4/21) Infected by Swine Influenza Virus (H1N1 and H3N2)
Abstract
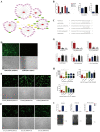
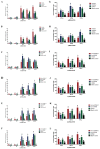
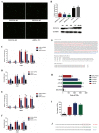
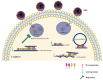
H1N1 and H3N2 are the two most common subtypes of swine influenza virus (SIV). They not only endanger the pig industry, but are also a huge risk of zoonotic diseases. However, the molecular mechanism and regulatory network of pigs (hosts) against influenza virus infection are still unclear. In this study, porcine alveolar macrophage cell (3D4/21) models infected by swine influenza virus (H1N1 and H3N2) were constructed. The expression profiles of miRNAs, mRNAs, lncRNAs and circRNAs after H1N1 and H3N2 infected 3D4/21 cells were revealed in this study. Then, two ceRNAs (TCONS_00166432-miR10391-MAN2A1 and novel_circ_0004733-miR10391-MAN2A1) that regulated H1N1 and H3N2 infection in 3D4/21 cells were verified by the methods of bioinformatics analysis, gene overexpression, gene interference, real-time quantitative PCR (qPCR), dual luciferase activity assay and RNA immunoprecipitation (RIP). In addition, the important candidate molecules (miR-10391, TCONS_00166432, and novel_circ_0004733) were identified by qPCR and enzyme linked immunosorbent assay (ELISA). Finally, the regulatory effect and possible molecular mechanism of the target gene MAN2A1 were identified by the methods of gene interference, qPCR, Western blot and ELISA. The results of this study suggested that TCONS_00166432 and novel_circ_0004733 could competitively bind miR-10391 to target the MAN2A1 gene to regulate swine influenza virus infecting 3D4/21 cells. This study reported for the first time the ceRNA networks involved in the regulation of the swine influenza virus infecting 3D4/21 cells, which provided a new insight into the molecular mechanism of 3D4/21 cells against swine influenza virus infection.
Keywords: ceRNA regulation; influenza virus; pig; porcine alveolar macrophage cell; whole transcriptome sequencing.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
Figures


References
-
- Sun Y., Ma S., Wei W., Xiao A., Xu Y., Dong J., Wang X. Epidemiological characteristics, diagnosis and prevention measures of swine influenza virus. Swine Ind. Sci. 2014;31:94–95. (In Chinese)
-
- Wang L., Bi Y., Cao Y., Li H. Isolation, identification and biological characterization of swine influenza virus Guangdong isolates. Chin. J. Vet. Sci. Technol. 2003;2:17–21. (In Chinese)
-
- Sun H., Xiao Y., Liu J., Wang D., Li F., Wang C., Li C., Zhu J., Song J., Sun H., et al. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc. Natl. Acad. Sci. USA. 2020;117:17204–17210. doi: 10.1073/pnas.1921186117. - DOI - PMC - PubMed
-
- Dhakal S., Hiremath J., Bondra K., Lakshmanappa Y.S., Shyu D., Ouyang K., Kang K., Binjawadagi B., Goodman J., Tabynov K., et al. Biodegradable nanoparticle delivery of inactivated swine influenza virus vaccine provides heterologous cell-mediated immune response in pigs. J. Control. Release. 2017;247:194–205. doi: 10.1016/j.jconrel.2016.12.039. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

