ERAP2 Inhibition Induces Cell-Surface Presentation by MOLT-4 Leukemia Cancer Cells of Many Novel and Potentially Antigenic Peptides
- PMID: 35163832
- PMCID: PMC8836666
- DOI: 10.3390/ijms23031913
ERAP2 Inhibition Induces Cell-Surface Presentation by MOLT-4 Leukemia Cancer Cells of Many Novel and Potentially Antigenic Peptides
Abstract
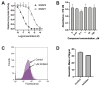
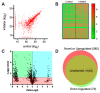
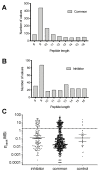
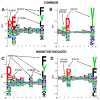
Recent studies have linked the activity of ER aminopeptidase 2 (ERAP2) to increased efficacy of immune-checkpoint inhibitor cancer immunotherapy, suggesting that pharmacological inhibition of ERAP2 could have important therapeutic implications. To explore the effects of ERAP2 inhibition on the immunopeptidome of cancer cells, we treated MOLT-4 T lymphoblast leukemia cells with a recently developed selective ERAP2 inhibitor, isolated Major Histocompatibility class I molecules (MHCI), and sequenced bound peptides by liquid chromatography tandem mass spectrometry. Inhibitor treatment induced significant shifts on the immunopeptidome so that more than 20% of detected peptides were either novel or significantly upregulated. Most of the inhibitor-induced peptides were 9mers and had sequence motifs and predicted affinity consistent with being optimal ligands for at least one of the MHCI alleles carried by MOLT-4 cells. Such inhibitor-induced peptides could serve as triggers for novel cytotoxic responses against cancer cells and synergize with the therapeutic effect of immune-checkpoint inhibitors.
Keywords: adaptive immunity; aminopeptidase; antigen presentation; antigenic peptide; immunopeptidome; inhibitor; major histocompatibility molecules; proteomics.
Conflict of interest statement
The authors declare that they have no competing financial interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

