Insights into Structure and Biological Activity of Copper(II) and Zinc(II) Complexes with Triazolopyrimidine Ligands
- PMID: 35164029
- PMCID: PMC8838430
- DOI: 10.3390/molecules27030765
Insights into Structure and Biological Activity of Copper(II) and Zinc(II) Complexes with Triazolopyrimidine Ligands
Abstract
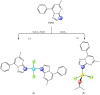
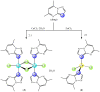
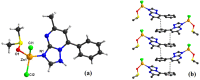
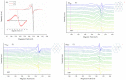
In an attempt to increase the biological activity of the 1,2,4-triazolo[1,5-a]pyrimidine scaffold through complexation with essential metal ions, the complexes trans-[Cu(mptp)2Cl2] (1), [Zn(mptp)Cl2(DMSO)] (2) (mptp: 5-methyl-7-phenyl-1,2,4-triazolo[1,5-a]pyrimidine), [Cu2(dmtp)4Cl4]·2H2O (3) and [Zn(dmtp)2Cl2] (4) (dmtp: 5,7-dimethyl-1,2,4-triazolo[1,5-a]pyrimidine), were synthesized and characterized as new antiproliferative and antimicrobial species. Both complexes (1) and (2) crystallize in the P21/n monoclinic space group, with the tetrahedral surroundings generating a square-planar stereochemistry in the Cu(II) complex and a tetrahedral stereochemistry in the Zn(II) species. The mononuclear units are interconnected in a supramolecular network through π-π interactions between the pyrimidine moiety and the phenyl ring in (1) while supramolecular chains resulting from C-H∙∙∙π interactions were observed in (2). All complexes exhibit an antiproliferative effect against B16 tumor cells and improved antibacterial and antifungal activities compared to the free ligands. Complex (3) displays the best antimicrobial activity against all four tested strains, both in the planktonic and biofilm-embedded states, which can be correlated to its stronger DNA-binding and nuclease-activity traits.
Keywords: 1,2,4-triazolo[1,5-a]pyrimidine; biofilm; complex; cytotoxicity; metallonuclease activity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Synthesis, Characterization, and in Vitro Anticancer Activity of Copper and Zinc Bis(Thiosemicarbazone) Complexes.Inorg Chem. 2019 Oct 21;58(20):13709-13723. doi: 10.1021/acs.inorgchem.9b01281. Epub 2019 Jul 24. Inorg Chem. 2019. PMID: 31339305
-
Zinc, cadmium, and mercury complexes of 5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidine (dmtp). Preparation, spectroscopic characterization, and biological activity. X-ray crystal structures of [Zn(dmtp)2Br2], [Hg(dmtp)2Cl2], and [Hdmtp]2[CdBr4]. 2H2O.J Inorg Biochem. 1996 Dec;64(4):259-71. doi: 10.1016/s0162-0134(96)00054-2. J Inorg Biochem. 1996. PMID: 8916414
-
Biological Activity of Triazolopyrimidine Copper(II) Complexes Modulated by an Auxiliary N-N-Chelating Heterocycle Ligands.Molecules. 2021 Nov 9;26(22):6772. doi: 10.3390/molecules26226772. Molecules. 2021. PMID: 34833864 Free PMC article.
-
Antibacterial, antifungal and in vitro antileukaemia activity of metal complexes with thiosemicarbazones.J Cell Mol Med. 2015 Apr;19(4):865-78. doi: 10.1111/jcmm.12508. Epub 2015 Feb 24. J Cell Mol Med. 2015. PMID: 25708540 Free PMC article.
-
Copper and Antibiotics: Discovery, Modes of Action, and Opportunities for Medicinal Applications.Adv Microb Physiol. 2017;70:193-260. doi: 10.1016/bs.ampbs.2017.01.007. Epub 2017 Mar 18. Adv Microb Physiol. 2017. PMID: 28528648 Review.
References
-
- Peng K., Liang B.-B., Liu W., Mao Z.-W. What blocks more anticancer platinum complexes from experiment to clinic: Major problems and potential strategies from drug design perspectives. Coord. Chem. Rev. 2021;449:214210. doi: 10.1016/j.ccr.2021.214210. - DOI
-
- Bertini I., Grey H.B., Stiefel E.I., Valentine J.S., editors. Biological Inorganic Chemistry. Structure and Reactivity. University Science Books; Sausalito, CA, USA: 2007.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

