Length-Dependent Deep Learning Model for RNA Secondary Structure Prediction
- PMID: 35164295
- PMCID: PMC8838716
- DOI: 10.3390/molecules27031030
Length-Dependent Deep Learning Model for RNA Secondary Structure Prediction
Abstract
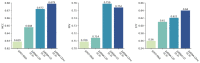
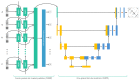
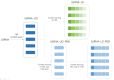
Deep learning methods for RNA secondary structure prediction have shown higher performance than traditional methods, but there is still much room to improve. It is known that the lengths of RNAs are very different, as are their secondary structures. However, the current deep learning methods all use length-independent models, so it is difficult for these models to learn very different secondary structures. Here, we propose a length-dependent model that is obtained by further training the length-independent model for different length ranges of RNAs through transfer learning. 2dRNA, a coupled deep learning neural network for RNA secondary structure prediction, is used to do this. Benchmarking shows that the length-dependent model performs better than the usual length-independent model.
Keywords: RNA secondary structure; deep learning; length-dependent model.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Predicting RNA secondary structure via adaptive deep recurrent neural networks with energy-based filter.BMC Bioinformatics. 2019 Dec 24;20(Suppl 25):684. doi: 10.1186/s12859-019-3258-7. BMC Bioinformatics. 2019. PMID: 31874602 Free PMC article.
-
RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning.Nat Commun. 2019 Nov 27;10(1):5407. doi: 10.1038/s41467-019-13395-9. Nat Commun. 2019. PMID: 31776342 Free PMC article.
-
sincFold: end-to-end learning of short- and long-range interactions in RNA secondary structure.Brief Bioinform. 2024 May 23;25(4):bbae271. doi: 10.1093/bib/bbae271. Brief Bioinform. 2024. PMID: 38855913 Free PMC article.
-
RNA structure prediction using deep learning - A comprehensive review.Comput Biol Med. 2025 Apr;188:109845. doi: 10.1016/j.compbiomed.2025.109845. Epub 2025 Feb 20. Comput Biol Med. 2025. PMID: 39983363 Review.
-
Revolutions in RNA secondary structure prediction.J Mol Biol. 2006 Jun 9;359(3):526-32. doi: 10.1016/j.jmb.2006.01.067. Epub 2006 Feb 6. J Mol Biol. 2006. PMID: 16500677 Review.
Cited by
-
Machine learning in RNA structure prediction: Advances and challenges.Biophys J. 2024 Sep 3;123(17):2647-2657. doi: 10.1016/j.bpj.2024.01.026. Epub 2024 Jan 30. Biophys J. 2024. PMID: 38297836 Review.
-
mRNA vaccine sequence and structure design and optimization: Advances and challenges.J Biol Chem. 2025 Jan;301(1):108015. doi: 10.1016/j.jbc.2024.108015. Epub 2024 Nov 26. J Biol Chem. 2025. PMID: 39608721 Free PMC article. Review.
-
Benchmarking the methods for predicting base pairs in RNA-RNA interactions.Bioinformatics. 2025 Jun 2;41(6):btaf289. doi: 10.1093/bioinformatics/btaf289. Bioinformatics. 2025. PMID: 40327448 Free PMC article.
-
Robust RNA secondary structure prediction with a mixture of deep learning and physics-based experts.Biol Methods Protoc. 2025 Jan 6;10(1):bpae097. doi: 10.1093/biomethods/bpae097. eCollection 2025. Biol Methods Protoc. 2025. PMID: 39811444 Free PMC article.
-
Examples of Structural Motifs in Viral Genomes and Approaches for RNA Structure Characterization.Int J Mol Sci. 2022 Dec 14;23(24):15917. doi: 10.3390/ijms232415917. Int J Mol Sci. 2022. PMID: 36555559 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

