CRP-Like Transcriptional Regulator MrpC Curbs c-di-GMP and 3',3'-cGAMP Nucleotide Levels during Development in Myxococcus xanthus
- PMID: 35164555
- PMCID: PMC8844925
- DOI: 10.1128/mbio.00044-22
CRP-Like Transcriptional Regulator MrpC Curbs c-di-GMP and 3',3'-cGAMP Nucleotide Levels during Development in Myxococcus xanthus
Abstract
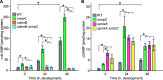
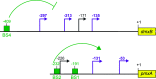
Myxococcus xanthus has a nutrient-regulated biphasic life cycle forming predatory swarms in the presence of nutrients and spore-filled fruiting bodies in the absence of nutrients. The second messenger 3'-5', 3'-5 cyclic di-GMP (c-di-GMP) is essential during both stages of the life cycle; however, different enzymes involved in c-di-GMP synthesis and degradation as well as several c-di-GMP receptors are important during distinct life cycle stages. To address this stage specificity, we determined transcript levels using transcriptome sequencing (RNA-seq) and transcription start sites using Cappable sequencing (Cappable-seq) during growth and development genome wide. All 70 genes encoding c-di-GMP-associated proteins were expressed, with 28 upregulated and 10 downregulated during development. Specifically, the three genes encoding enzymatically active proteins with a stage-specific function were expressed stage specifically. By combining operon mapping with published chromatin immunoprecipitation sequencing (ChIP-seq) data for MrpC (M. Robinson, B. Son, D. Kroos, L. Kroos, BMC Genomics 15:1123, 2014, http://dx.doi.org/10.1186/1471-2164-15-1123), the cAMP receptor protein (CRP)-like master regulator of development, we identified nine developmentally regulated genes as regulated by MrpC. In particular, MrpC directly represses the expression of dmxB, which encodes the diguanylate cyclase DmxB that is essential for development and responsible for the c-di-GMP increase during development. Moreover, MrpC directly activates the transcription of pmxA, which encodes a bifunctional phosphodiesterase that degrades c-di-GMP and 3',3'-cGAMP in vitro and is essential for development. Thereby, MrpC regulates and curbs the cellular pools of c-di-GMP and 3',3'-cGAMP during development. We conclude that temporal regulation of the synthesis of proteins involved in c-di-GMP metabolism contributes to c-di-GMP signaling specificity. MrpC is important for this regulation, thereby being a key regulator of developmental cyclic di-nucleotide metabolism in M. xanthus. IMPORTANCE The second messenger c-di-GMP is important during both stages of the nutrient-regulated biphasic life cycle of Myxococcus xanthus with the formation of predatory swarms in the presence of nutrients and spore-filled fruiting bodies in the absence of nutrients. However, different enzymes involved in c-di-GMP synthesis and degradation are important during distinct life cycle stages. Here, we show that the three genes encoding enzymatically active proteins with a stage-specific function are expressed stage specifically. Moreover, we find that the master transcriptional regulator of development MrpC directly regulates the expression of dmxB, which encodes the diguanylate cyclase DmxB that is essential for development, and of pmxA, which encodes a bifunctional phosphodiesterase that degrades c-di-GMP and 3',3'-cGAMP in vitro and is essential for development. We conclude that temporal regulation of the synthesis of proteins involved in c-di-GMP metabolism contributes to c-di-GMP signaling specificity and that MrpC plays an important role in this regulation.
Keywords: 3'; 3'-cGAMP; CRP; CRP-like proteins; Cappable-seq; PilZ; c-di-GMP; cGAMP; cyclic nucleotides; development; diguanylate cyclase; fruiting body formation; phosphodiesterase; second messenger; sporulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
