Isolation of a Novel Bat Rhabdovirus with Evidence of Human Exposure in China
- PMID: 35164557
- PMCID: PMC8844929
- DOI: 10.1128/mbio.02875-21
Isolation of a Novel Bat Rhabdovirus with Evidence of Human Exposure in China
Abstract
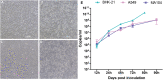
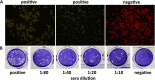
Bats are well-recognized reservoirs of zoonotic viruses. Several spillover events from bats to humans have been reported, causing severe epidemic or endemic diseases including severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), SARS-CoV, Middle East respiratory syndrome-CoV (MERS-CoV), henipaviruses, and filoviruses. In this study, a novel rhabdovirus species, provisionally named Rhinolophus rhabdovirus DPuer (DPRV), was identified from the horseshoe bat (Rhinolophus affinis) in Yunnan province, China, using next-generation sequencing. DPRV shedding in the spleen, liver, lung, and intestinal contents of wild bats with high viral loads was detected by real-time quantitative PCR, indicating that DPRV has tropism for multiple host tissues. Furthermore, DPRV can replicate in vitro in multiple mammalian cell lines, including BHK-21, A549, and MA104 cells, with the highest efficiency in hamster kidney cell line BHK-21, suggesting infectivity of DPRV in these cell line-derived hosts. Ultrastructure analysis revealed a characteristic bullet-shaped morphology and tightly clustered distribution of DPRV particles in the intracellular space. DPRV replicated efficiently in suckling mouse brains and caused death of suckling mice; death rates increased with passaging of DPRV in suckling mice. Moreover, 421 serum samples were collected from individuals who lived near the bat collection site and had fever symptoms within 1 year. DPRV-specific antibodies were detected in 20 (4.75%) human serum samples by indirect immunofluorescence assay. Furthermore, 10 (2.38%) serum samples were DPRV positive according to plaque reduction neutralization assay, which revealed potential transmission of DPRV from bats to humans and highlighted the potential public health risk. Potential vector association with DPRV was not found with negative viral RNA in bloodsucking arthropods. IMPORTANCE We identified a novel rhabdovirus from the horseshoe bat (Rhinolophus thomasi) in China with probable infectivity in humans. DPRV was isolated in vitro from several mammalian cell lines, indicating wide host tropism, excluding bats, of DPRV. DPRV replicated in the brains of suckling mice, and the death rate of suckling mice increased with passaging of DPRV in vivo. Serological tests indicated the possible infectivity of DPRV in humans and the potential transmission to humans. The present findings provide preliminary evidence for the potential risk of DPRV to public health. Additional studies with active surveillance are needed to address interspecies transmission and determine the pathogenicity of DPRV in humans.
Keywords: Ledantevirus; bats; cell culture; complete genome; pathogenicity; phylogeny; rhabdovirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Broad Cross-Species Infection of Cultured Cells by Bat HKU2-Related Swine Acute Diarrhea Syndrome Coronavirus and Identification of Its Replication in Murine Dendritic Cells In Vivo Highlight Its Potential for Diverse Interspecies Transmission.J Virol. 2019 Nov 26;93(24):e01448-19. doi: 10.1128/JVI.01448-19. Print 2019 Dec 15. J Virol. 2019. PMID: 31554686 Free PMC article.
-
Epidemiology and Genomic Characterization of Two Novel SARS-Related Coronaviruses in Horseshoe Bats from Guangdong, China.mBio. 2022 Jun 28;13(3):e0046322. doi: 10.1128/mbio.00463-22. Epub 2022 Apr 25. mBio. 2022. PMID: 35467426 Free PMC article.
-
Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination.J Virol. 2015 Oct;89(20):10532-47. doi: 10.1128/JVI.01048-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269185 Free PMC article.
-
Molecular epidemiology, evolution and phylogeny of SARS coronavirus.Infect Genet Evol. 2019 Jul;71:21-30. doi: 10.1016/j.meegid.2019.03.001. Epub 2019 Mar 4. Infect Genet Evol. 2019. PMID: 30844511 Free PMC article. Review.
-
Emerging viruses: Cross-species transmission of coronaviruses, filoviruses, henipaviruses, and rotaviruses from bats.Cell Rep. 2022 Jun 14;39(11):110969. doi: 10.1016/j.celrep.2022.110969. Epub 2022 May 30. Cell Rep. 2022. PMID: 35679864 Free PMC article. Review.
Cited by
-
Bifidobacteria define gut microbiome profiles of golden lion tamarin (Leontopithecus rosalia) and marmoset (Callithrix sp.) metagenomic shotgun pools.Sci Rep. 2023 Sep 21;13(1):15679. doi: 10.1038/s41598-023-42059-4. Sci Rep. 2023. PMID: 37735195 Free PMC article.
-
Treponema denticola major surface protein (Msp): a key player in periodontal pathogenicity and immune evasion.Arch Microbiol. 2025 Jan 18;207(2):36. doi: 10.1007/s00203-024-04223-w. Arch Microbiol. 2025. PMID: 39825920 Review.
-
Broad geographical circulation of a novel vesiculovirus in bats in the Mediterranean region.PLoS Negl Trop Dis. 2025 Jun 12;19(6):e0013172. doi: 10.1371/journal.pntd.0013172. eCollection 2025 Jun. PLoS Negl Trop Dis. 2025. PMID: 40504866 Free PMC article.
References
-
- Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, Bryden WL, Middleton D, Reid PA, McFarlane RA, Martin G, Tabor GM, Skerratt LF, Anderson DL, Crameri G, Quammen D, Jordan D, Freeman P, Wang L-F, Epstein JH, Marsh GA, Kung NY, McCallum H. 2015. Ecological dynamics of emerging bat virus spillover. Proc Biol Sci 282:20142124. doi:10.1098/rspb.2014.2124. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

