Inhibition of Protein N- Glycosylation Blocks SARS-CoV-2 Infection
- PMID: 35164559
- PMCID: PMC8844921
- DOI: 10.1128/mbio.03718-21
Inhibition of Protein N- Glycosylation Blocks SARS-CoV-2 Infection
Abstract
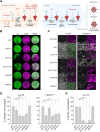
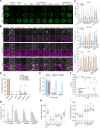
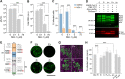
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) extensively N-glycosylates its spike proteins, which are necessary for host cell invasion and the target of both vaccines and immunotherapies. These N-glycans are predicted to modulate spike binding to the host receptor by stabilizing its open conformation and host immunity evasion. Here, we investigated the essentiality of both the host N-glycosylation pathway and SARS-CoV-2 N-glycans for infection. Ablation of host N-glycosylation using RNA interference or inhibitors, including FDA-approved drugs, reduced the spread of the infection, including that of variants B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma) and B.1.617.2 (Delta). Under these conditions, cells produced fewer virions and some completely lost their infectivity. Furthermore, partial enzymatic deglycosylation of intact virions showed that surface-exposed N-glycans are critical for cell invasion. Altogether, we propose protein N-glycosylation as a targetable pathway with clinical potential for treatment of COVID-19. IMPORTANCE The coronavirus SARS-CoV-2 uses its spike surface proteins to infect human cells. Spike proteins are heavily modified with several N-glycans, which are predicted to modulate their function. In this work, we show that interfering with either the synthesis or attachment of spike N-glycans significantly reduces the spread of SARS-CoV-2 infection in vitro, including that of several variants. As new SARS-CoV-2 variants, with various degrees of resistance against current vaccines, are likely to continue appearing, halting virus glycosylation using repurposed human drugs could result in a complementary strategy to reducing the spread of COVID-19 worldwide.
Keywords: COVID-19; N-glycosylation; SARS-CoV-2; antiviral agents; coronavirus; glycosylation; viral infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. doi: 10.1016/S0140-6736(20)30251-8. - DOI - PMC - PubMed
-
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

