MonaGO: a novel gene ontology enrichment analysis visualisation system
- PMID: 35164667
- PMCID: PMC8845231
- DOI: 10.1186/s12859-022-04594-1
MonaGO: a novel gene ontology enrichment analysis visualisation system
Abstract
Background: Gene ontology (GO) enrichment analysis is frequently undertaken during exploration of various -omics data sets. Despite the wide array of tools available to biologists to perform this analysis, meaningful visualisation of the overrepresented GO in a manner which is easy to interpret is still lacking.
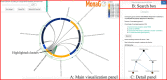
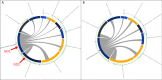
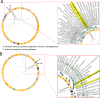
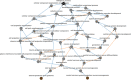
Results: Monash Gene Ontology (MonaGO) is a novel web-based visualisation system that provides an intuitive, interactive and responsive interface for performing GO enrichment analysis and visualising the results. MonaGO supports gene lists as well as GO terms as inputs. Visualisation results can be exported as high-resolution images or restored in new sessions, allowing reproducibility of the analysis. An extensive comparison between MonaGO and 11 state-of-the-art GO enrichment visualisation tools based on 9 features revealed that MonaGO is a unique platform that simultaneously allows interactive visualisation within one single output page, directly accessible through a web browser with customisable display options.
Conclusion: MonaGO combines dynamic clustering and interactive visualisation as well as customisation options to assist biologists in obtaining meaningful representation of overrepresented GO terms, producing simplified outputs in an unbiased manner. MonaGO will facilitate the interpretation of GO analysis and will assist the biologists into the representation of the results.
Keywords: GO enrichment; Gene ontology; Interactive visualisation; Semantic web; Web services.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

