Degeneracy measures in biologically plausible random Boolean networks
- PMID: 35164672
- PMCID: PMC8845291
- DOI: 10.1186/s12859-022-04601-5
Degeneracy measures in biologically plausible random Boolean networks
Abstract
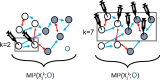
Background: Degeneracy-the ability of structurally different elements to perform similar functions-is a property of many biological systems. Highly degenerate systems show resilience to perturbations and damage because the system can compensate for compromised function due to reconfiguration of the underlying network dynamics. Degeneracy thus suggests how biological systems can thrive despite changes to internal and external demands. Although degeneracy is a feature of network topologies and seems to be implicated in a wide variety of biological processes, research on degeneracy in biological networks is mostly limited to weighted networks. In this study, we test an information theoretic definition of degeneracy on random Boolean networks, frequently used to model gene regulatory networks. Random Boolean networks are discrete dynamical systems with binary connectivity and thus, these networks are well-suited for tracing information flow and the causal effects. By generating networks with random binary wiring diagrams, we test the effects of systematic lesioning of connections and perturbations of the network nodes on the degeneracy measure.
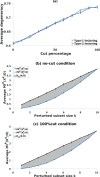
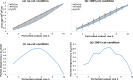
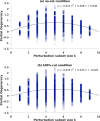
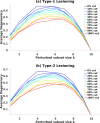
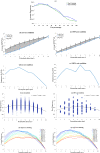
Results: Our analysis shows that degeneracy, on average, is the highest in networks in which ~ 20% of the connections are lesioned while 50% of the nodes are perturbed. Moreover, our results for the networks with no lesions and the fully-lesioned networks are comparable to the degeneracy measures from weighted networks, thus we show that the degeneracy measure is applicable to different networks.
Conclusions: Such a generalized applicability implies that degeneracy measures may be a useful tool for investigating a wide range of biological networks and, therefore, can be used to make predictions about the variety of systems' ability to recover function.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Sadowski MI, Jones DT. The sequence–structure relationship and protein function prediction. Curr Opin Struct Biol. 2009;19(3):357–362. - PubMed
-
- Pal D, Eisenberg D. Inference of protein function from protein structure. Structure. 2005;13(1):121–130. - PubMed
-
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(6761):C47–52. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

