The POR rs10954732 polymorphism decreases susceptibility to hepatocellular carcinoma and hepsin as a prognostic biomarker correlated with immune infiltration based on proteomics
- PMID: 35164791
- PMCID: PMC8842912
- DOI: 10.1186/s12967-022-03282-1
The POR rs10954732 polymorphism decreases susceptibility to hepatocellular carcinoma and hepsin as a prognostic biomarker correlated with immune infiltration based on proteomics
Abstract
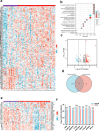
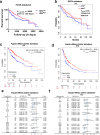
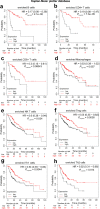
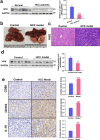
The effect of the cytochrome P450 oxidoreductase (POR) rs10954732 (G > A) polymorphism on hepatocellular carcinoma (HCC) susceptibility is unknown. Here we found that A allele carriers showed a 69% decrease in susceptibility to HCC with overall survival (OS) prolonged to 199%, accompanied by lower activity for cytochrome P450 2E1. A total of 222 differentially expressed proteins were mainly enriched in neutrophil and T cell activation and involved in the immune and inflammatory responses, constituting the altered immune tumor microenvironment related with A allele by proteomics analysis. Hepsin (HPN) showed significant down-regulation in HCC and up-regulation in A allele carriers. A lower HPN level was associated with increased susceptibility to HCC and a worse prognosis. Moreover, HPN is a potential independent prognostic biomarker for HCC and is strongly associated with clinicopathological features, tumor-infiltrating status of immune cells both in our discovery cohort and database surveys. Our findings provide a new potential mechanism by which HPN may play an important role in the susceptibility of rs10954732 A allele carriers to HCC and their prognosis through tumor immune infiltration, thus offering potential insights for future studies on tumor immunotherapy.
Keywords: HPN; Hepatocellular carcinoma; POR; Polymorphisms; Proteomics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures




Similar articles
-
Use of proteomics to identify mechanisms of hepatocellular carcinoma with the CYP2D6*10 polymorphism and identification of ANGPTL6 as a new diagnostic and prognostic biomarker.J Transl Med. 2021 Aug 19;19(1):359. doi: 10.1186/s12967-021-03038-3. J Transl Med. 2021. PMID: 34412629 Free PMC article.
-
Prognostic analysis of tumor mutation burden and immune infiltration in hepatocellular carcinoma based on TCGA data.Aging (Albany NY). 2021 Apr 4;13(8):11257-11280. doi: 10.18632/aging.202811. Epub 2021 Apr 4. Aging (Albany NY). 2021. PMID: 33820866 Free PMC article.
-
Transcriptional analysis of the expression, prognostic value and immune infiltration activities of the COMMD protein family in hepatocellular carcinoma.BMC Cancer. 2021 Sep 7;21(1):1001. doi: 10.1186/s12885-021-08699-3. BMC Cancer. 2021. PMID: 34493238 Free PMC article.
-
CXCL2/10/12/14 are prognostic biomarkers and correlated with immune infiltration in hepatocellular carcinoma.Biosci Rep. 2021 Jun 25;41(6):BSR20204312. doi: 10.1042/BSR20204312. Biosci Rep. 2021. PMID: 34085699 Free PMC article.
-
CDKN2A is a prognostic biomarker and correlated with immune infiltrates in hepatocellular carcinoma.Biosci Rep. 2021 Oct 29;41(10):BSR20211103. doi: 10.1042/BSR20211103. Biosci Rep. 2021. PMID: 34405225 Free PMC article.
Cited by
-
KIAA1429 facilitates progression of hepatocellular carcinoma by modulating m6A levels in HPN.Heliyon. 2023 Nov 8;9(11):e22084. doi: 10.1016/j.heliyon.2023.e22084. eCollection 2023 Nov. Heliyon. 2023. PMID: 38058614 Free PMC article.
-
DNA methylation in peripheral blood leukocytes for the association with glucose metabolism and invasive breast cancer.Clin Epigenetics. 2023 Feb 13;15(1):23. doi: 10.1186/s13148-023-01435-7. Clin Epigenetics. 2023. PMID: 36782224 Free PMC article.
-
Human Cytochrome P450 Cancer-Related Metabolic Activities and Gene Polymorphisms: A Review.Cells. 2024 Nov 26;13(23):1958. doi: 10.3390/cells13231958. Cells. 2024. PMID: 39682707 Free PMC article. Review.
-
Analysis of the Relationship Between Parkinson's Disease and Diabetic Retinopathy Based on Bioinformatics Methods.Mol Neurobiol. 2024 Sep;61(9):6395-6406. doi: 10.1007/s12035-024-03982-3. Epub 2024 Feb 3. Mol Neurobiol. 2024. PMID: 38308666
-
Proteomic analysis of DEN and CCl4-induced hepatocellular carcinoma mouse model.Sci Rep. 2024 Apr 5;14(1):8013. doi: 10.1038/s41598-024-58587-6. Sci Rep. 2024. PMID: 38580754 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

