Patient-specific Boolean models of signalling networks guide personalised treatments
- PMID: 35164900
- PMCID: PMC9018074
- DOI: 10.7554/eLife.72626
Patient-specific Boolean models of signalling networks guide personalised treatments
Abstract
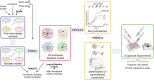
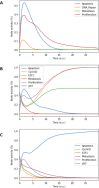
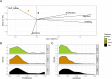
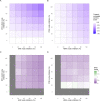
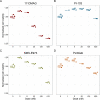
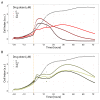
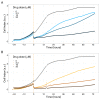
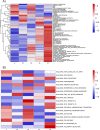
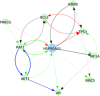
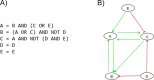
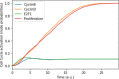
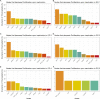
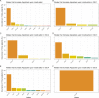
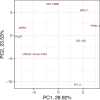
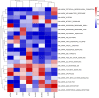
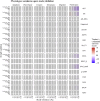
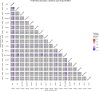
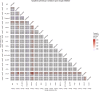
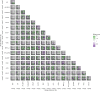
Prostate cancer is the second most occurring cancer in men worldwide. To better understand the mechanisms of tumorigenesis and possible treatment responses, we developed a mathematical model of prostate cancer which considers the major signalling pathways known to be deregulated. We personalised this Boolean model to molecular data to reflect the heterogeneity and specific response to perturbations of cancer patients. A total of 488 prostate samples were used to build patient-specific models and compared to available clinical data. Additionally, eight prostate cell line-specific models were built to validate our approach with dose-response data of several drugs. The effects of single and combined drugs were tested in these models under different growth conditions. We identified 15 actionable points of interventions in one cell line-specific model whose inactivation hinders tumorigenesis. To validate these results, we tested nine small molecule inhibitors of five of those putative targets and found a dose-dependent effect on four of them, notably those targeting HSP90 and PI3K. These results highlight the predictive power of our personalised Boolean models and illustrate how they can be used for precision oncology.
Keywords: computational biology; drug combinations; human; logical modelling; personalised drug; personalised medicine; prostate cancer; simulations; systems biology.
© 2022, Montagud et al.
Conflict of interest statement
AM, JB, PT, BS, EB, LC No competing interests declared, LT is a full-time employee and shareholder of AstraZeneca, VS is a full-time employee of AstraZeneca, RA is CEO of Astridbio Technologies Ltd, LP is a scientific advisor of Astridbio Technologies Ltd, AV Reviewing editor, eLife, JS reports funding from GSK and Sanofi and fees from Travere Therapeutics and Astex
Figures















References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

