Targeting the Ubiquitin-Proteasome System Using the UBA1 Inhibitor TAK-243 is a Potential Therapeutic Strategy for Small-Cell Lung Cancer
- PMID: 35165102
- PMCID: PMC9365348
- DOI: 10.1158/1078-0432.CCR-21-0344
Targeting the Ubiquitin-Proteasome System Using the UBA1 Inhibitor TAK-243 is a Potential Therapeutic Strategy for Small-Cell Lung Cancer
Abstract
Purpose: Small cell lung cancer (SCLC) is an aggressive disease with an overall 5-year survival rate of less than 10%. Treatment for SCLC with cisplatin/etoposide chemotherapy (C/E) ± radiotherapy has changed modestly over several decades. The ubiquitin-proteasome system is an underexplored therapeutic target for SCLC. We preclinically evaluated TAK-243, a first-in-class small molecule E1 inhibitor against UBA1.
Experimental design: We assessed TAK-243 in 26 SCLC cell-lines as monotherapy and combined with C/E, the PARP-inhibitor, olaparib, and with radiation using cell viability assays. We interrogated TAK-243 response with gene expression to identify candidate biomarkers. We evaluated TAK-243 alone and in combination with olaparib or radiotherapy with SCLC patient-derived xenografts (PDX).
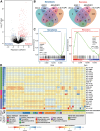
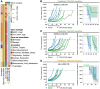
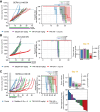
Results: Most SCLC cell lines were sensitive to TAK-243 monotherapy (EC50 median 15.8 nmol/L; range 10.2 nmol/L-367.3 nmol/L). TAK-243 sensitivity was associated with gene-sets involving the cell cycle, DNA and chromatin organization, and DNA damage repair, while resistance associated with cellular respiration, translation, and neurodevelopment. These associations were also observed in SCLC PDXs. TAK-243 synergized with C/E and olaparib in vitro across sensitive and resistant SCLC cell lines. Considerable TAK-243-olaparib synergy was observed in an SCLC PDX resistant to both drugs individually. TAK-243 radiosensitization was also observed in an SCLC PDX.
Conclusions: TAK-243 displays efficacy in SCLC preclinical models. Enrichment of gene sets is associated with TAK-243 sensitivity and resistance. TAK-243 exhibits synergy when combined with genotoxic therapies in cell lines and PDXs. TAK-243 is a potential therapeutic strategy to improve SCLC patient outcomes, both as a single agent and in combination with existing therapies.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures


Comment in
-
New potential targeted strategies in small cell lung cancer.Transl Lung Cancer Res. 2023 Apr 28;12(4):661-664. doi: 10.21037/tlcr-23-64. Epub 2023 Apr 4. Transl Lung Cancer Res. 2023. PMID: 37197617 Free PMC article. No abstract available.
References
-
- Simpson KL, Stoney R, Frese KK, Simms N, Rowe W, Pearce SP, et al. A biobank of small cell lung cancer CDX models elucidates inter- and intratumoral phenotypic heterogeneity. Nat Cancer 2020;1:437–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

