Structural basis of neuropeptide Y signaling through Y1 receptor
- PMID: 35165283
- PMCID: PMC8844075
- DOI: 10.1038/s41467-022-28510-6
Structural basis of neuropeptide Y signaling through Y1 receptor
Erratum in
-
Author Correction: Structural basis of neuropeptide Y signaling through Y1 receptor.Nat Commun. 2022 Feb 25;13(1):1126. doi: 10.1038/s41467-022-28837-0. Nat Commun. 2022. PMID: 35217654 Free PMC article. No abstract available.
Abstract
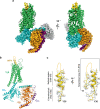
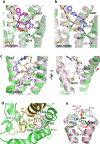
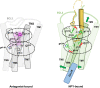
Neuropeptide Y (NPY) is highly abundant in the brain and involved in various physiological processes related to food intake and anxiety, as well as human diseases such as obesity and cancer. However, the molecular details of the interactions between NPY and its receptors are poorly understood. Here, we report a cryo-electron microscopy structure of the NPY-bound neuropeptide Y1 receptor (Y1R) in complex with Gi1 protein. The NPY C-terminal segment forming the extended conformation binds deep into the Y1R transmembrane core, where the amidated C-terminal residue Y36 of NPY is located at the base of the ligand-binding pocket. Furthermore, the helical region and two N-terminal residues of NPY interact with Y1R extracellular loops, contributing to the high affinity of NPY for Y1R. The structural analysis of NPY-bound Y1R and mutagenesis studies provide molecular insights into the activation mechanism of Y1R upon NPY binding.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Michel MC, et al. XVI. International Union of Pharmacology recommendations for the nomenclature of neuropeptide Y, peptide YY, and pancreatic polypeptide receptors. Pharm. Rev. 1998;50:143–150. - PubMed
-
- Kohno D, Yada T. Arcuate NPY neurons sense and integrate peripheral metabolic signals to control feeding. Neuropeptides. 2012;46:315–319. - PubMed
-
- Gotzsche CR, Woldbye DP. The role of NPY in learning and memory. Neuropeptides. 2016;55:79–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

