Bioinformatic and experimental characterization of SEN1998: a conserved gene carried by the Enterobacteriaceae-associated ROD21-like family of genomic islands
- PMID: 35165310
- PMCID: PMC8844411
- DOI: 10.1038/s41598-022-06183-x
Bioinformatic and experimental characterization of SEN1998: a conserved gene carried by the Enterobacteriaceae-associated ROD21-like family of genomic islands
Abstract
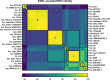
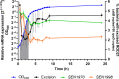
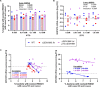
Genomic islands (GIs) are horizontally transferred elements that shape bacterial genomes and contributes to the adaptation to different environments. Some GIs encode an integrase and a recombination directionality factor (RDF), which are the molecular GI-encoded machinery that promotes the island excision from the chromosome, the first step for the spread of GIs by horizontal transfer. Although less studied, this process can also play a role in the virulence of bacterial pathogens. While the excision of GIs is thought to be similar to that observed in bacteriophages, this mechanism has been only studied in a few families of islands. Here, we aimed to gain a better understanding of the factors involved in the excision of ROD21 a pathogenicity island of the food-borne pathogen Salmonella enterica serovar Enteritidis and the most studied member of the recently described Enterobacteriaceae-associated ROD21-like family of GIs. Using bioinformatic and experimental approaches, we characterized the conserved gene SEN1998, showing that it encodes a protein with the features of an RDF that binds to the regulatory regions involved in the excision of ROD21. While deletion or overexpression of SEN1998 did not alter the expression of the integrase-encoding gene SEN1970, a slight but significant trend was observed in the excision of the island. Surprisingly, we found that the expression of both genes, SEN1998 and SEN1970, were negatively correlated to the excision of ROD21 which showed a growth phase-dependent pattern. Our findings contribute to the growing body of knowledge regarding the excision of GIs, providing insights about ROD21 and the recently described EARL family of genomic islands.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Marcoleta AE, Berríos-Pastén C, Nuñez G, Monasterio O, Lagos R. Klebsiella pneumoniae asparagine tDNAs are integration hotspots for different genomic islands encoding microcin E492 production determinants and other putative virulence factors present in hypervirulent strains. Front. Microbiol. 2016;7:849. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

