Stain-free detection of embryo polarization using deep learning
- PMID: 35165311
- PMCID: PMC8844381
- DOI: 10.1038/s41598-022-05990-6
Stain-free detection of embryo polarization using deep learning
Abstract
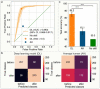
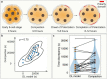
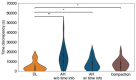
Polarization of the mammalian embryo at the right developmental time is critical for its development to term and would be valuable in assessing the potential of human embryos. However, tracking polarization requires invasive fluorescence staining, impermissible in the in vitro fertilization clinic. Here, we report the use of artificial intelligence to detect polarization from unstained time-lapse movies of mouse embryos. We assembled a dataset of bright-field movie frames from 8-cell-stage embryos, side-by-side with corresponding images of fluorescent markers of cell polarization. We then used an ensemble learning model to detect whether any bright-field frame showed an embryo before or after onset of polarization. Our resulting model has an accuracy of 85% for detecting polarization, significantly outperforming human volunteers trained on the same data (61% accuracy). We discovered that our self-learning model focuses upon the angle between cells as one known cue for compaction, which precedes polarization, but it outperforms the use of this cue alone. By compressing three-dimensional time-lapsed image data into two-dimensions, we are able to reduce data to an easily manageable size for deep learning processing. In conclusion, we describe a method for detecting a key developmental feature of embryo development that avoids clinically impermissible fluorescence staining.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Label free cell-tracking and division detection based on 2D time-lapse images for lineage analysis of early embryo development.Comput Biol Med. 2014 Aug;51:24-34. doi: 10.1016/j.compbiomed.2014.04.011. Epub 2014 May 9. Comput Biol Med. 2014. PMID: 24873887 Free PMC article.
-
Delay of polarization event increases the number of Cdx2-positive blastomeres in mouse embryo.Dev Biol. 2012 Aug 1;368(1):54-62. doi: 10.1016/j.ydbio.2012.05.013. Epub 2012 May 17. Dev Biol. 2012. PMID: 22609553
-
Automated embryo stage classification in time-lapse microscopy video of early human embryo development.Med Image Comput Comput Assist Interv. 2013;16(Pt 2):460-7. doi: 10.1007/978-3-642-40763-5_57. Med Image Comput Comput Assist Interv. 2013. PMID: 24579173
-
Polarity in Cell-Fate Acquisition in the Early Mouse Embryo.Curr Top Dev Biol. 2016;120:203-34. doi: 10.1016/bs.ctdb.2016.04.008. Epub 2016 Jun 7. Curr Top Dev Biol. 2016. PMID: 27475853 Review.
-
Mouse Embryo Compaction.Curr Top Dev Biol. 2016;120:235-58. doi: 10.1016/bs.ctdb.2016.04.005. Epub 2016 May 17. Curr Top Dev Biol. 2016. PMID: 27475854 Review.
Cited by
-
Combined Input Deep Learning Pipeline for Embryo Selection for In Vitro Fertilization Using Light Microscopic Images and Additional Features.J Imaging. 2025 Jan 7;11(1):13. doi: 10.3390/jimaging11010013. J Imaging. 2025. PMID: 39852326 Free PMC article.
-
From beasts to bytes: Revolutionizing zoological research with artificial intelligence.Zool Res. 2023 Nov 18;44(6):1115-1131. doi: 10.24272/j.issn.2095-8137.2023.263. Zool Res. 2023. PMID: 37933101 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

