LONP-1 and ATFS-1 sustain deleterious heteroplasmy by promoting mtDNA replication in dysfunctional mitochondria
- PMID: 35165413
- PMCID: PMC8868506
- DOI: 10.1038/s41556-021-00840-5
LONP-1 and ATFS-1 sustain deleterious heteroplasmy by promoting mtDNA replication in dysfunctional mitochondria
Abstract
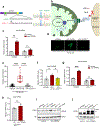
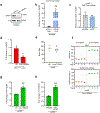
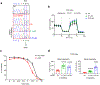
The accumulation of deleterious mitochondrial DNA (∆mtDNA) causes inherited mitochondrial diseases and ageing-associated decline in mitochondrial functions such as oxidative phosphorylation. Following mitochondrial perturbations, the bZIP protein ATFS-1 induces a transcriptional programme to restore mitochondrial function. Paradoxically, ATFS-1 is also required to maintain ∆mtDNAs in heteroplasmic worms. The mechanism by which ATFS-1 promotes ∆mtDNA accumulation relative to wild-type mtDNAs is unclear. Here we show that ATFS-1 accumulates in dysfunctional mitochondria. ATFS-1 is absent in healthy mitochondria owing to degradation by the mtDNA-bound protease LONP-1, which results in the nearly exclusive association between ATFS-1 and ∆mtDNAs in heteroplasmic worms. Moreover, we demonstrate that mitochondrial ATFS-1 promotes the binding of the mtDNA replicative polymerase (POLG) to ∆mtDNAs. Interestingly, inhibition of the mtDNA-bound protease LONP-1 increased ATFS-1 and POLG binding to wild-type mtDNAs. LONP-1 inhibition in Caenorhabditis elegans and human cybrid cells improved the heteroplasmy ratio and restored oxidative phosphorylation. Our findings suggest that ATFS-1 promotes mtDNA replication in dysfunctional mitochondria by promoting POLG-mtDNA binding, which is antagonized by LONP-1.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests Statement
The authors declare no competing interests.
Figures
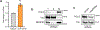



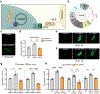
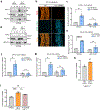
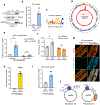




Comment in
-
LONP-1 aids propagation of deleterious mtDNA.Nat Cell Biol. 2022 Feb;24(2):127-128. doi: 10.1038/s41556-021-00838-z. Nat Cell Biol. 2022. PMID: 35165415 No abstract available.
References
-
- Baker BM & Haynes CM Mitochondrial protein quality control during biogenesis and aging. Trends in biochemical sciences 36, 254–261 (2011). - PubMed
-
- Wai T, Teoli D & Shoubridge EA The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nature genetics 40, 1484–1488 (2008). - PubMed
-
- Srivastava S & Moraes CT Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease. Human molecular genetics 10, 3093–3099 (2001). - PubMed
-
- Tanaka M et al. Gene therapy for mitochondrial disease by delivering restriction endonucleaseSmaI into mitochondria. Journal of biomedical science 9, 534–541 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

