Mitochondrial fission links ECM mechanotransduction to metabolic redox homeostasis and metastatic chemotherapy resistance
- PMID: 35165418
- PMCID: PMC7615745
- DOI: 10.1038/s41556-022-00843-w
Mitochondrial fission links ECM mechanotransduction to metabolic redox homeostasis and metastatic chemotherapy resistance
Abstract
Metastatic breast cancer cells disseminate to organs with a soft microenvironment. Whether and how the mechanical properties of the local tissue influence their response to treatment remains unclear. Here we found that a soft extracellular matrix empowers redox homeostasis. Cells cultured on a soft extracellular matrix display increased peri-mitochondrial F-actin, promoted by Spire1C and Arp2/3 nucleation factors, and increased DRP1- and MIEF1/2-dependent mitochondrial fission. Changes in mitochondrial dynamics lead to increased production of mitochondrial reactive oxygen species and activate the NRF2 antioxidant transcriptional response, including increased cystine uptake and glutathione metabolism. This retrograde response endows cells with resistance to oxidative stress and reactive oxygen species-dependent chemotherapy drugs. This is relevant in a mouse model of metastatic breast cancer cells dormant in the lung soft tissue, where inhibition of DRP1 and NRF2 restored cisplatin sensitivity and prevented disseminated cancer-cell awakening. We propose that targeting this mitochondrial dynamics- and redox-based mechanotransduction pathway could open avenues to prevent metastatic relapse.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The invention of Drpitor1a is the subject of a US Patent Application 20200323829 from D.W. and S.L.A. The remaining authors declare no competing interests.
Figures

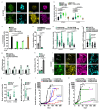
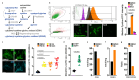



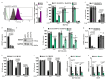


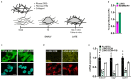
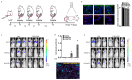

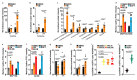



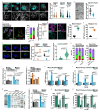

Comment in
-
Softening redox homeostasis in cancer cells.Nat Cell Biol. 2022 Feb;24(2):133-134. doi: 10.1038/s41556-022-00845-8. Nat Cell Biol. 2022. PMID: 35165419 Free PMC article.
References
-
- Petridou NI, Spiró Z, Heisenberg C-P. Multiscale force sensing in development. Nature Cell Biology. 2017;19:581–588. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

